On this page we will try to explain some behaviour of the match engines in our Search and Match application.
Antigen Recognition Domain explained
- The antigen recognition domain (ARD) is the binding groove of the HLA peptide. This is the region interacting with the presented antigen and T-cell receptor.
- All alleles that express the same amino acid sequence in this region are considered an allele match.
- The ARD is encoded on:
- exon 2 & 3 for HLA class I
- exon 2 for HLA class II
1. Null allele treatment
- Null alleles are treated as absent, i.e. the second typing matches as homozygous.
- Null allele matching rules are applied to:
- All high resolution null alleles.
- Null alleles as part of multi allele codes if the null allele is part of a haplotype matching the donor‘s (patient‘s) HLA typing.
- Null allele matching rules are only applied to one typing of the locus.
The following table explains situations where Hap-E performs null allele treatment and whether this is a considered a (potential) match or not.
Patient typing | Donor typing | Hap-E | ATLAS |
|---|---|---|---|
A*03:01, A*01:11N | A*03:01, A*03:01 | matches | matches |
A*03:01, A*01:11N | A*03:01, A*02:125N | matches | matches |
If there is a haplotype compatible with the donor typing containing A*02:125N then | |||
A*03:01, A*01:11N | A*03:01, A*02:GFJM | matches | mismatches |
A*03:01, A*03:01 | A*03:01, A*02:XX | matches | mismatch, but in list as if no mismatch |
A*03:01, A*03:01 | A*03:01, A*01:01:01G | does not match | does not match |
A*01:11N, A*02:125N | A*03:01, A*03:01 | does not match | does not match |
02:GFJM ≜ 02:01/02:105/02:125N
2. Search with two mismatches
Hap-E search | ATLAS search |
|---|---|
| The two mismatches can be on any locus | The two mismatches can be on any locus |
3. Donors with DNA and serologic typing
Hap-E search | ATLAS search |
|---|---|
Only the DNA typing information is used for matching | Only the DNA typing information is used for matching |
4. Serology handling
Hap-E search | ATLAS search |
|---|---|
Does not assign assumed serology or expert assigned exceptions described in rel_dna_ser e.g. Hap-E assigns A*;02:55 to A(2) | Does assign assumed serology or expert assigned exceptions described in rel_dna_ser e.g. ATLAS assigns A*;02:55 to A(2) and A(28) |
Split serology will only match same split serology or when donor/patient is broad serology. Not matched when patient/donor has split serology that is different but both share same broad serology. e.g.
| Split serology will matched when patient/donor has split serology that is different but both share same broad serology |
5. Returning donors without typing at DRB1
Hap-E search | ATLAS search |
|---|---|
Yes, when default 'return only donors with typing at DRB1' filter has been switched off and donor does have typing at A+B and at least 1 of: C, DQB1 loci | No, because donors without typing at DRB1 are not accepted by ATLAS as valid donors |
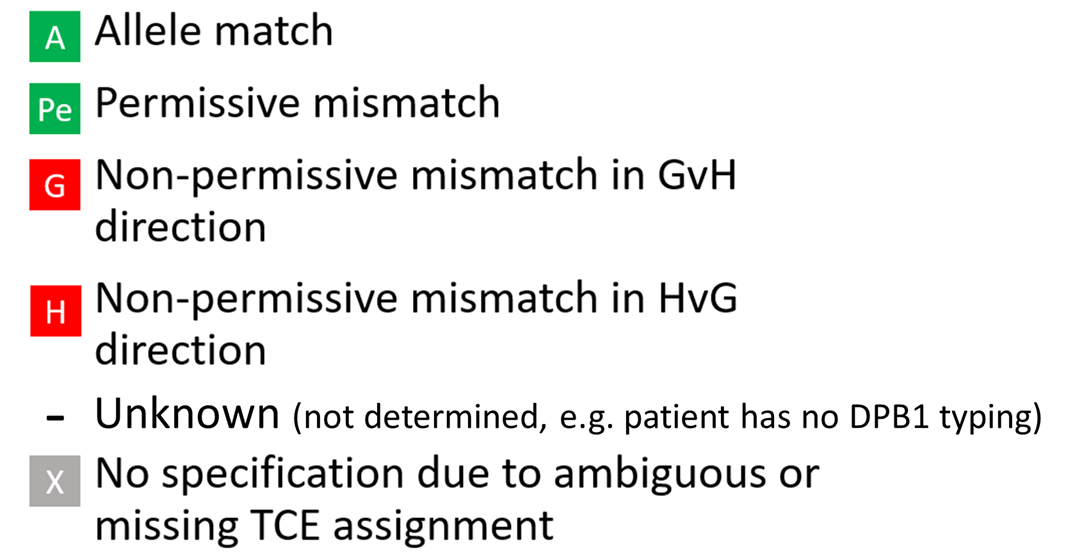
6. Match grades: Overview
Standard match grades |
|---|
Applied to locus A, B, C, DRB1, DQB1 |
7. DPB1-specific match grades
Hap-E search | ATLAS search |
|---|---|
Applied to DPB1 only. Uses TCE3 model version 2 | Applied to DPB1 only. Uses TCE3 model version 2 |
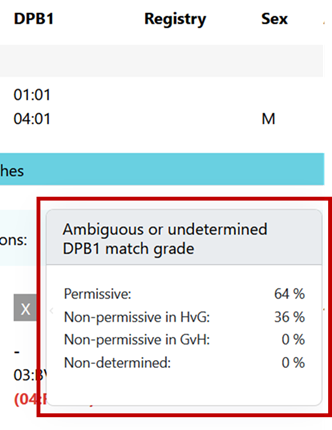
Ambiguous typing can be determined. For an ambiguous case (i.e. depending on the realization of a multi allele code on DPB1 of patient or donor, the mismatch can be P, H or G) the probabilities are calculated based on DPB1 allele frequencies and are displayed when hovering over the displayed DPB1 match-grade icon of the donor or CBU: | Ambiguous typing can't be determined. Ambiguous typing is displayed as "-" (Unknown) |
8. Handling of HLA*DRB3, DRB4, DRB5, DQA1 and other loci not considered for overall match probability
Hap-E search | ATLAS search |
|---|---|
HLA*DRB3/4/5, DPA1, DQA1 and E are validated and shown in the full report, including information whether they are matched or not | Loci other than A, B, C, DRB1, DQB1 and DPB1 are not validated or imported by ATLAS and therefore also not returned |
9. Handling of 'NEW' alleles
Handling of new alleles that do not have an official name yet.
For guidelines regarding implementation and reporting of 'NEW' alleles, please read https://onlinelibrary.wiley.com/doi/10.1111/tan.15048
Hap-E search | ATLAS search |
|---|---|
|
|
|
|
10. Cord Blood search options
Hap-E search | ATLAS search |
|---|---|
Allele matched:
| Allele matched:
|
|
11. 3 and 4 mismatch Cord Blood sorting
Hap-E search | ATLAS search |
|---|---|
Resolution based scoring CBUs with high resolution typing are ranked higher than less well typed CBUs in the 3 and 4 mismatch category e.g. DNA typed CBUs are ranked higher than serology typed CBUs | Match confidence based scoring CBUs with a higher match confidence are ranked higher than CBUs with a lower match confidence in the 3 and 4 mismatch category e.g. CBUs with exact matches at considered loci are ranked higher than those with possible matches at considered loci |
12. Probability display
Hap-E search | ATLAS search |
|---|---|
All probabilities are rounded to integer values → 0%:
| All probabilities are rounded to integer values → 0%:
|
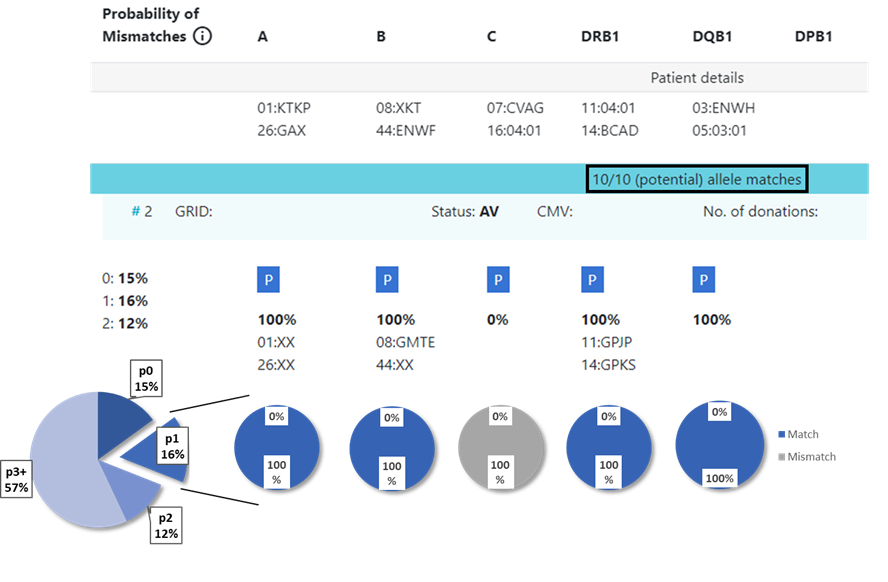
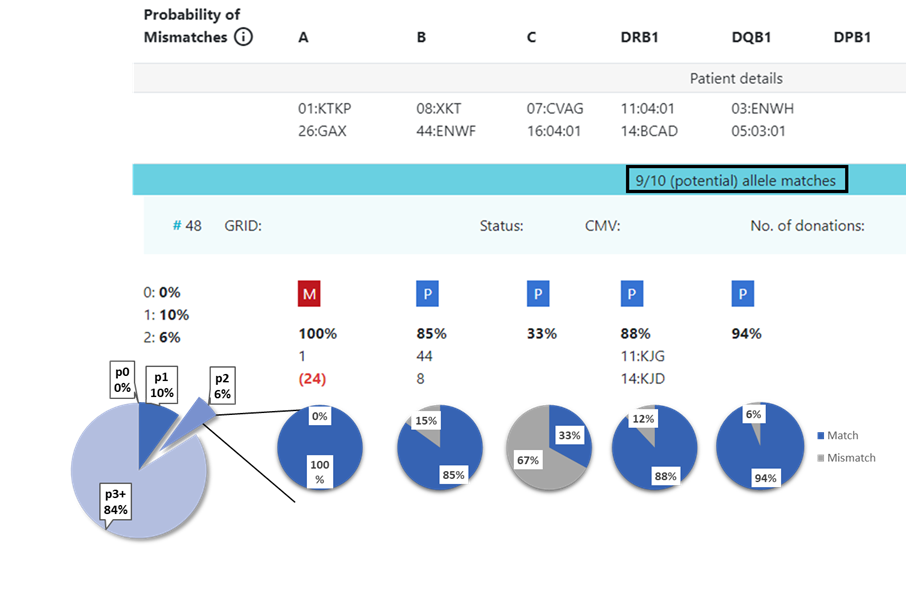
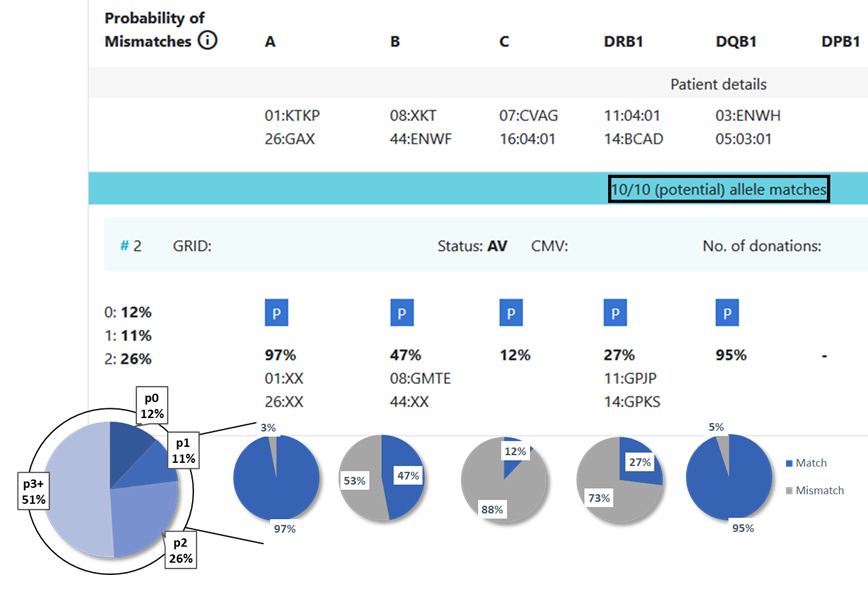
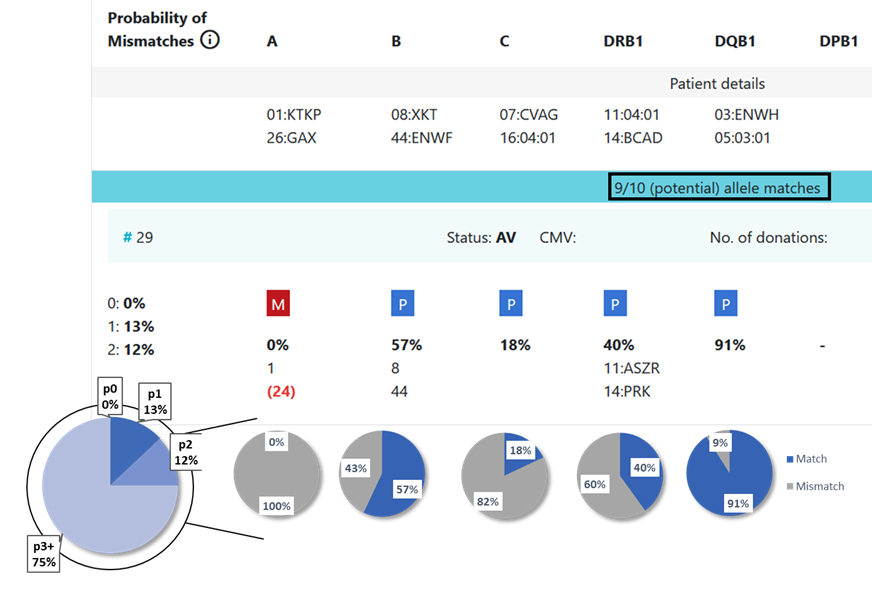
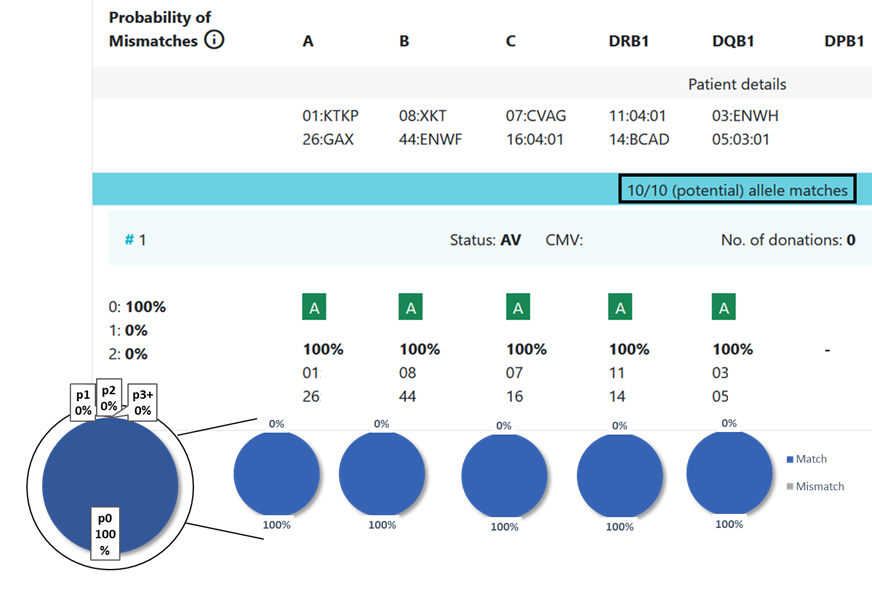
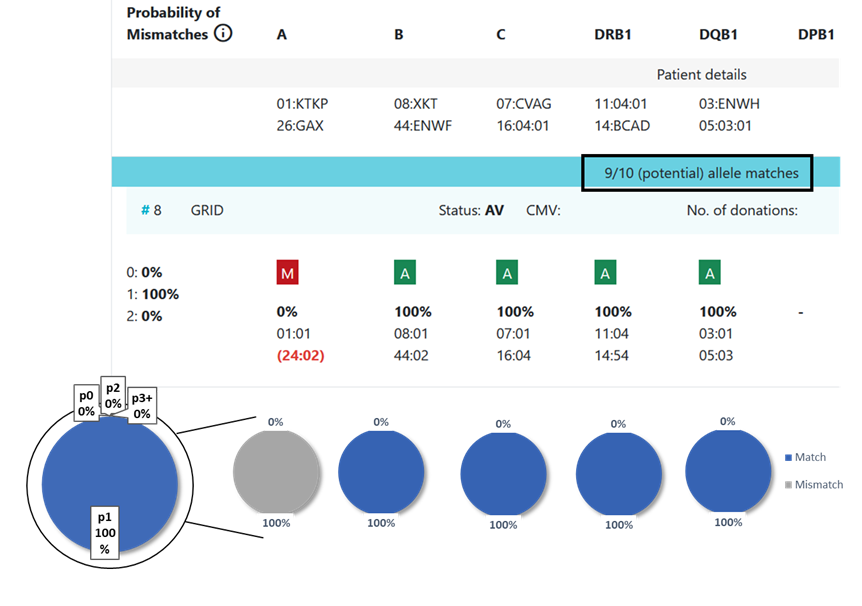
13.1 Locus match probabilities: Definition
Hap-E search | ATLAS search |
|---|---|
match, i.e. 10/10, 8/8 or 6/6
mismatch, i.e. 9/10, 7/8 or 5/6
So this probability provides information on which locus the next mismatch will occur | match, i.e. 10/10, 8/8 or 6/6
mismatch, i.e. 9/10, 7/8 or 5/6
So this probability provides information on the probability that this locus will be a match |
13.2 Locus match probabilities in search: Not displayed
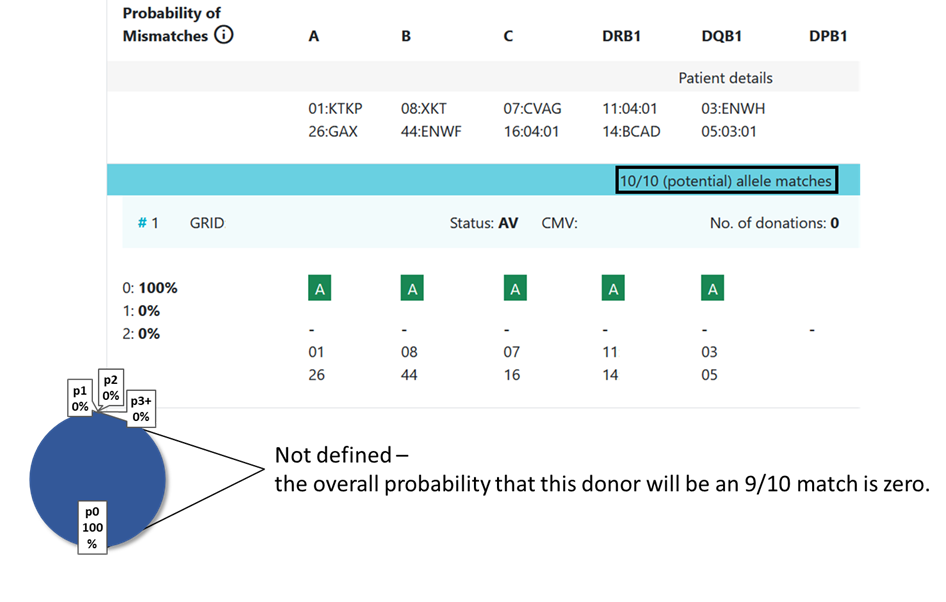
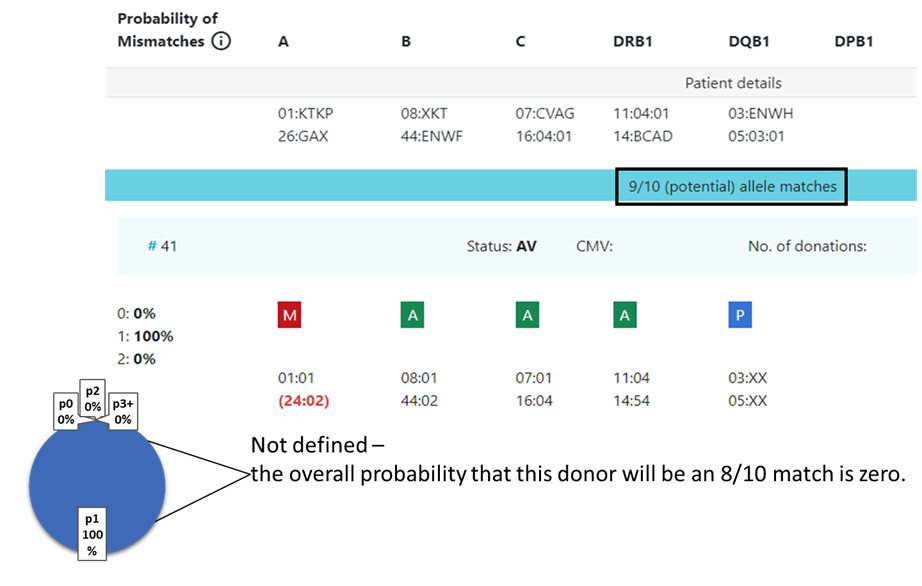
In some cases the value is not defined and no locus match probability is displayed:
Hap-E search | ATLAS search |
|---|---|
match
| match
mismatch
inexplicable donors
|