...
| Expand | ||
|---|---|---|
| ||
Patients for whom a search was performed within the last month 6 weeks will be in the ‘Active patient’ list, while the previous 5 months will be in the ‘Inactive patient’ list. If you cannot find your patient within the default Active list remember to search the Inactive list also. Users will have access to patients registered by other users at their organisation as well, which is also split into 'Active Patients' and 'Inactive patients' |
...
| Expand | ||
|---|---|---|
| ||
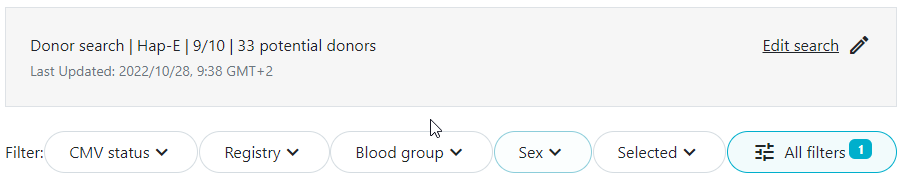
Hap-E searches may return donors that do not have typing at DRB1, but do have typing at loci A, B and at least one other locus. Since these donors are usually not considered relevant potential donors and there tend to be quite a few of these donors that potentially match the patient, Search & Match applies a default filter which removes all donors that do not have typing at DRB1. You can see this by the little "1" next to the filter. This means that there is already a filter applied to the results. If you are interested in the donors without DRB1 typing you can remove the filter and see all results. |
| Expand | ||
|---|---|---|
| ||
Asynchronous matching is a process of running searches in parallel. It allows you to enter a patient and start the search run, and proceed with other activities within the application like entering another patient or reviewing another match list while the search runs in the background. You do not have to wait for the current search to finish before proceeding with other activities. |
| Expand | ||
|---|---|---|
| ||
Many factors can impact how long it takes to retrieve search results. The most likely reasons that your search is taking some time is that it is a difficult search or the HLA of your patient is very common and will retrieve many records. We initially only run a 10/10, 8/8 or 6/6 search to make sure we retrieve you results as quickly as possible. Mismatch searches can take longer since there are many more options to assess before providing the results. |
| Expand | ||
|---|---|---|
| ||
| It is not necessary to cancel a search. We recommend logging out of the application and returning at a later time to review your results. |
| Expand | ||
|---|---|---|
| ||
By default, Search & Match performs a 10/10 search for donors if all 5 loci (A,B,C,DRB1,DQB1) are available for the patient. If only 3 or 4 loci are available, the system will scale down to a 6/6 or 8/8 search, respectively. 10/10, 8/8 and 6/6 are all considered as zero mismatch searches and will not include mismatch donors in the results. This is also the case for cord searches. |
| Expand | ||
|---|---|---|
| ||
Open up the search results for a search you would like to perform a mismatch search. Click "Edit search" and then select the number of mismatches that you would like to consider at most. Then click "Search" to start that search. |
| Expand | ||
|---|---|---|
| ||
No, you can only perform a zero mismatch donor or cords search. You will only be able to view/request mismatched donors/CBUs when the initial search results are available. |
| Expand | ||
|---|---|---|
| ||
| WMDA has calculated and applied many specific haplotype frequency sets to the Search & Match Service. Read more on the following page. |
| Expand | ||
|---|---|---|
| ||
If you are experiencing difficulties in your selection of a donor/cord for your patient, you can request a search advice from the WMDA HLA experts. To request an advise, go to your patient list, click on the patient ID and at the bottom of the Update patient form you will find a button with 'Request Search Advisory'. An email message will then open with the patient information and some additional fields with requirements. Please fill out as much as possible and send the email. You will receive the advice also by email. |
| Expand | ||
|---|---|---|
| ||
There is no functional difference. The filters you see on top of the headings in the search results are the frequently used filters. All filters are applied to all search results. |
...
| title | It seems like the search results are automatically ordered on age for adults and TNC for cords. Is that correct? |
|---|
The default sorting criteria from Hap-E Search and ATLAS for donors are:
1. HLA
2. probability in 10% intervals
3. donor age ascending
The default sorting criteria from Hap-E Search and ATLAS for cords are:
1. HLA (6/6, 5/6, 4/6 categories)
2. Number of total nucleated cells (TNC) for cords within HLA match category
...
| title | What are the DBP1 TC3 evaluation conditions |
|---|
| |
If this occurs the most likely cause is:
The reason this donor/CBU is shown as a potential match and not a mismatch is that the typing for the donor contains a null allele. It is therefore a possibility that the donor actually has a null allele. Since a homozygous typing is generally considered a match with a donor that has one allele that matches the homozygous patient typing and the other is a null allele, Hap-E returns this donor/CBU as a potential match. Please see the slide below from a recent webinar: |
| Expand | ||
|---|---|---|
| ||
In the legacy system when running a standard donor search, all returned donors have values for A, B and DRB1. This is different when running a search in the new system. There all donors need to have typing at A, B and at least one other locus (which may or may not be DRB1). This means that donors are returned in the new system that do not have typing at DRB1. This typically explains the majority of the difference in number of search results as mentioned in the search summary. Although in some cases these donors with typing at A, B and C or DQB1 may prove useful, in most cases they are not. We therefore apply a default filter to all search results. You can see all donors by unchecking the default filter. Because the filter is applied by default, you may notice a difference in the total number of donors/cords as described in the search summary text versus the number displayed just above the list of potential matches. For example in the example below, there are 33 donors in the full search results. 3 donors did not have typing at DRB1 and have therefore been filtered out in the donor search results shown. You can see that a filter has been applied by the "1" icon on the "All filters" button. |
| Expand | ||
|---|---|---|
| ||
Asynchronous matching is a process of running searches in parallel. It allows you to enter a patient and start the search run, and proceed with other activities within the application like entering another patient or reviewing another match list while the search runs in the background. You do not have to wait for the current search to finish before proceeding with other activities. |
| Expand | ||
|---|---|---|
| ||
Many factors can impact how long it takes to retrieve search results. The most likely reasons that your search is taking some time is that it is a difficult search or the HLA of your patient is very common and will retrieve many records. We initially only run a 10/10, 8/8 or 6/6 search to make sure we retrieve you results as quickly as possible. Mismatch searches can take longer since there are many more options to assess before providing the results. |
| Expand | ||
|---|---|---|
| ||
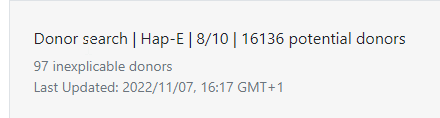
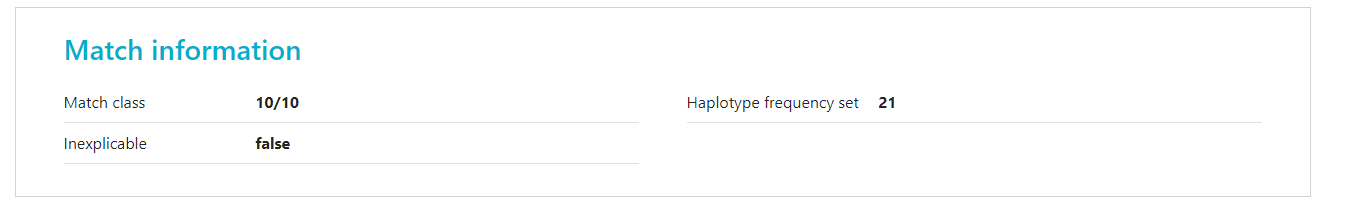
Inexplicable donors or CBUs are records that have HLA that cannot be explained by a combination of two haplotypes in the haplotype frequency (HF) set for the population that the donor or CBU is part of. Because the HLA typing of this donor/CBU cannot be explained, the matching algorithm is unable to calculate match probabilities. Currently these donors appear on the bottom of the match class that they are part of (e.g. 10/10, 9/10, 7/8). The number of inexplicable donors/CBUs mentioned therefore serves as a reminder that there are potentially relevant search results available that can be found at the bottom of the match class. |
| Expand | ||
|---|---|---|
| ||
| It is not necessary to cancel a search. We recommend logging out of the application and returning at a later time to review your results. |
| Expand | ||
|---|---|---|
| ||
By default, Search & Match performs a 10/10 search for donors if all 5 loci (A,B,C,DRB1,DQB1) are available for the patient. If only 3 or 4 loci are available, the system will scale down to a 6/6 or 8/8 search, respectively. 10/10, 8/8 and 6/6 are all considered as zero mismatch searches and will not include mismatch donors in the results. This is also the case for cord searches. |
| Expand | ||
|---|---|---|
| ||
Open up the search results for a search you would like to perform a mismatch search. Click "Edit search" and then select the number of mismatches that you would like to consider at most. Then click "Search" to start that search. |
| Expand | ||
|---|---|---|
| ||
No, you can only perform a zero mismatch donor or cords search. You will only be able to view/request mismatched donors/CBUs when the initial search results are available. |
| Expand | ||
|---|---|---|
| ||
| WMDA has calculated and applied many specific haplotype frequency sets to the Search & Match Service. Read more on the following page. To find which haplotype frequency set got used for a donor or CBU, consult the full report. |
| Expand | ||
|---|---|---|
| ||
If you are experiencing difficulties in your selection of a donor/cord for your patient, you can request a search advice from the WMDA HLA experts. To request an advise, go to your patient list, click on the patient ID and at the bottom of the Update patient form you will find a button with 'Request Search Advisory'. An email message will then open with the patient information and some additional fields with requirements. Please fill out as much as possible and send the email. You will receive the advice also by email. |
| Expand | ||
|---|---|---|
| ||
There is no functional difference. The filters you see on top of the headings in the search results are the frequently used filters. All filters are applied to all search results. Even the ones that are not on the page that is currently displayed. |
| Expand | ||
|---|---|---|
| ||
The default sorting criteria from Hap-E Search and ATLAS for donors are: 1. HLA 2. probability in 10% intervals 3. donor age ascending The default sorting criteria from Hap-E Search and ATLAS for cords are: 1. HLA (6/6, 5/6, 4/6 categories) 2. Number of total nucleated cells (TNC) for cords within HLA match category |
| Expand | ||
|---|---|---|
| ||
DPB1 TCE3 evaluation is performed only for A-B-DR(B1) typed donors under the following conditions:
As part of the DPB1 TC3 grading filter, you can filter the results by the following options: Matched, Permissive, Non-permissive in HvG direction, Non-permissive in GvH direction, Ambiguous/Undetermined. Although the DPB1 TC3 filter is available for cords, the results haven't been validated so it shouldn't be used. |
| Expand | ||
|---|---|---|
| ||
The default sorting criteria from Hap-E Search and ATLAS are: 1. HLA 2. probability in 10% intervals 3. donor age in 5 year intervals |
| Expand | ||
|---|---|---|
| ||
This can have several causes.
See the following page for more information: Feature differences Hap-E Search vs Optimatch |
| Expand | ||
|---|---|---|
| ||
In some cases you may see donors or CBUs that look like the one above. There is not information regarding GRID, registry, CBB, age or any other non-HLA/ethnicity information. The reason for this is that the search was in the Hap-E search result, but can no longer be found in our internal database. We are therefore not able to show any additional information other then what the Hap-E matching engine provided. This can have several causes.
How to resolve:
|
| Expand | ||
|---|---|---|
| ||
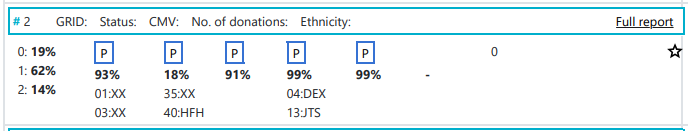
At the top of search results you can see how many donors or CBUs are inexplicable. Inexplicable donors or CBUs are records that have HLA that cannot be explained by a combination of two haplotypes in the haplotype frequency set (Haplotype frequency sets in the HAP-E matching algorithm of the Search & Match Service) for the population that the donor or CBU is part of. To find out which haplotype frequency set was used, you can use the full report The number there corresponds to the set on the Haplotype frequency sets in the HAP-E matching algorithm of the Search & Match Service share page. Because the HLA typing that this donor/CBU has cannot be explained, Hap-E also cannot calculate match probabilities. Currently these donors appear on the bottom of the match class that they are part of (e.g. 10/10, 9/10, 7/8). We are working on a way to move potentially relevant but inexplicable donors to a better place in the search results. The number of inexplicable donors/CBUs mentioned therefore serves as a reminder that there are potentially relevant search results available that can be found at the bottom of the match class. |
DPB1 TCE3 evaluation is performed only for A-B-DR(B1) typed donors under the following conditions:
- Patient DPB1 values must be present. Typing ambiguities in the form of multiple alleles codes, G-codes, etc. are allowed.
- Donor DPB1 values must be present. Typing ambiguities in the form of multiple alleles codes, G-codes, etc. are allowed.
As part of the DPB1 TC3 grading filter, you can filter the results by the following options: Matched, Permissive, Non-permissive in HvG direction, Non-permissive in GvH direction, Ambiguous/Undetermined.
Although the DPB1 TC3 filter is available for cords, the results haven't been validated so it shouldn't be used.
| Expand | ||
|---|---|---|
| ||
The default sorting criteria from Hap-E Search and ATLAS are: 1. HLA 2. probability in 10% intervals 3. donor age in 5 year intervals |
| Expand | ||
|---|---|---|
| ||
This can have several causes.
See the following page for more information: Feature differences Hap-E Search vs Optimatch |
| Expand | ||
|---|---|---|
| ||
Since B*15:BPXE = B*15:03/61/74/103 a donor with this codes is a potential allele match for the patient. According to the official WMDA serology/DNA correspondence table, B*15:61 and B*15:74 have a serology of B15/B70 while B*15:03 is B72 and B*15:103 is B70. As a consequence a serology of B15 rules out B*15:03 and this donor is no longer potentially identical (on the allele level). Another explanation could be the limited length of donor lists. Background: Unfortunately, for many donors serology was derived from DNA (by using the first field for the serological assignment) and vice versa (by appending “:XX” to the serological assignment) and often eventually both values are reported. In the case discussed, B*15:BPXE probably was translated back into B15 which is most likely wrong. This is a typical and (with certain registries) frequent case but there are many more unexpected DNA-serology correspondences that can give rise to exactly the same situation. |
...