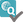...
This section includes your personalised dashboard, which shows information about the number of active patients from you and your organisation. It also presents the numbers of patients with an error, patients without searches performed (only registered), searches that are still running and the search reports that are available. Furthermore, you will find on this page some practical information, links to the addresses of registries and cord blood banks and information how to report feedback and problems. (figure 6: personal homepage)
Currently no home page
| Imagefloat | ||
|---|---|---|
| ||
...
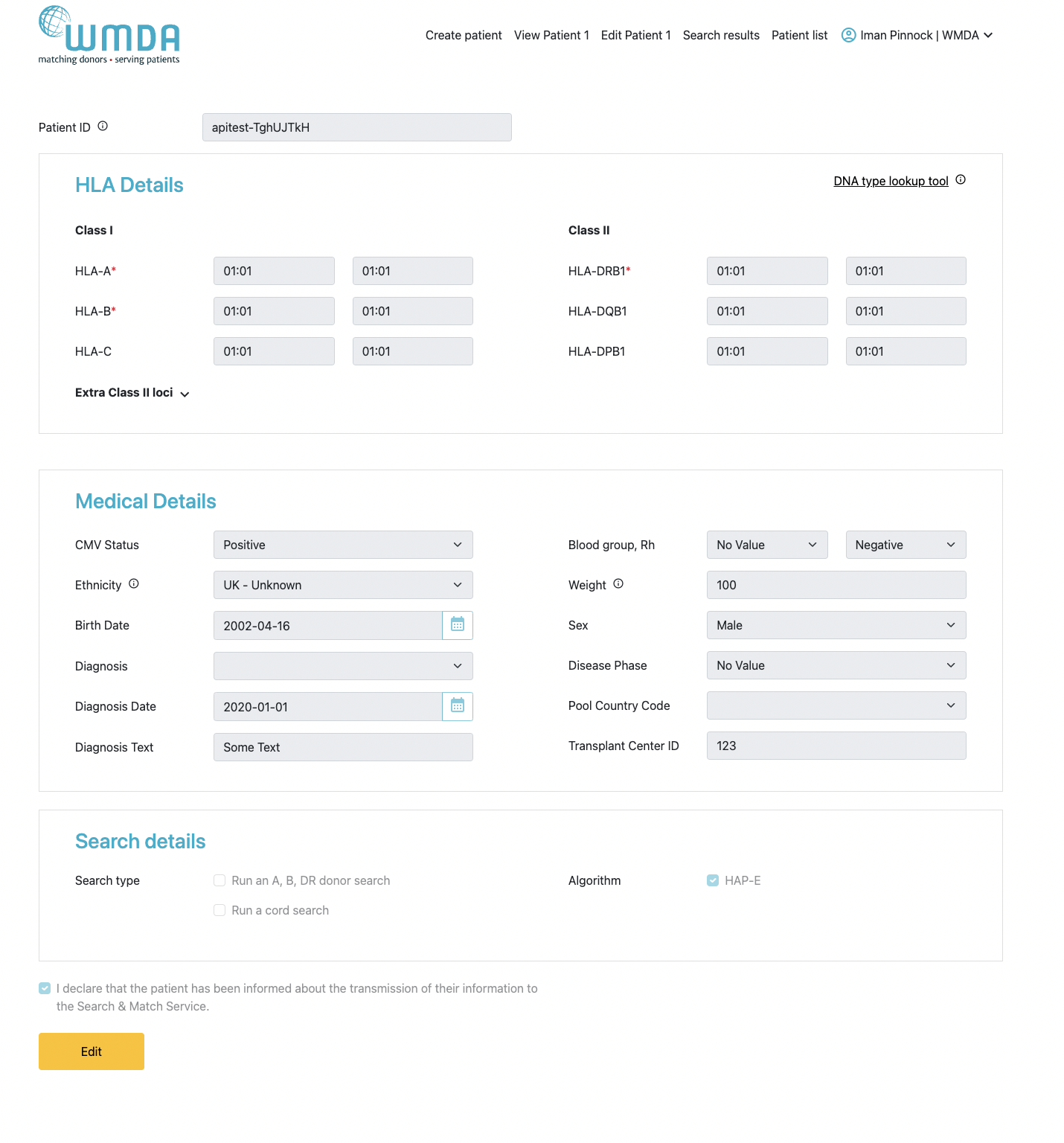
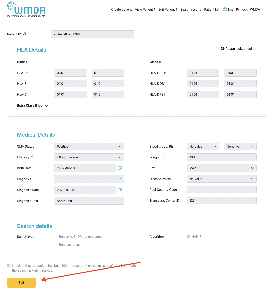
The "update patient" form is accessed by clicking on a patient in the patient list.
It additionally shows the match results if you already have performed a match run. The button "Add patient and run match" is replaced by the button "update patient and re-run match". Furthermore, 2 extra buttons are visible: "Request Search Advisory" (for requesting advice on difficult searches from the WMDA HLA expert) and "Deactivate patient" (for moving a patient from the active patient list to the inactive patient list).
When entering a Patient, only the following fields are mandatory:
...
| Imagefloat | ||
|---|---|---|
| ||
|
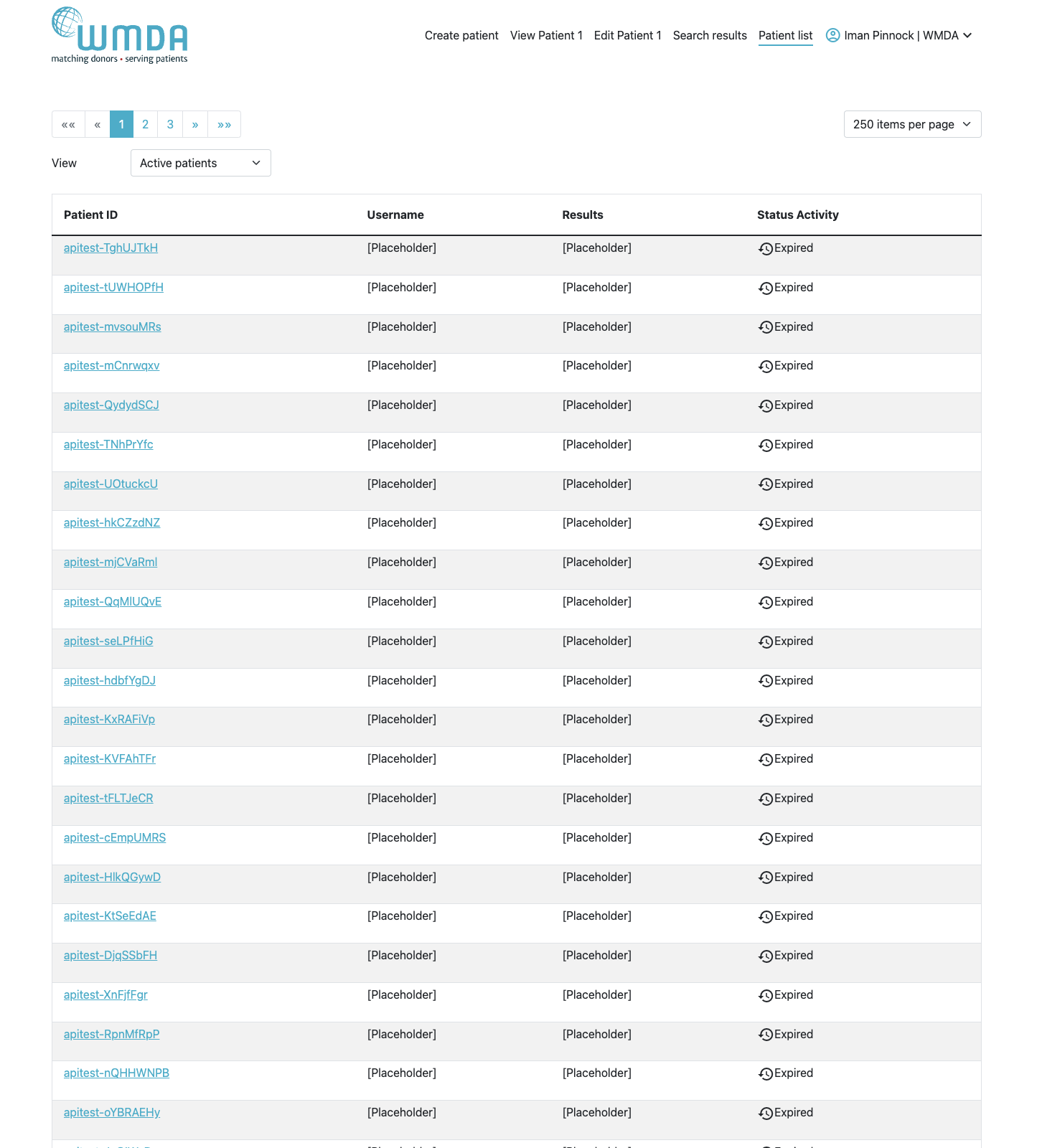
Within the patient list you can perform various other functions listed below:
...
Actions | Description |
Patient ID Link | Clicking on the Patient ID link will open the update patient form to allow users to perform the following functions: Edit/Update patient details: Modify any details for the given patient, except the Patient ID.
PLEASE NOTE: Update of any of the patients details such as HLA, will automatically trigger a new match run. This might change previous search results. Only if you remove all ticks in the search type block, no new match run is triggered, but any previous search results will be lost as well. |
(Search) Results Link |
|
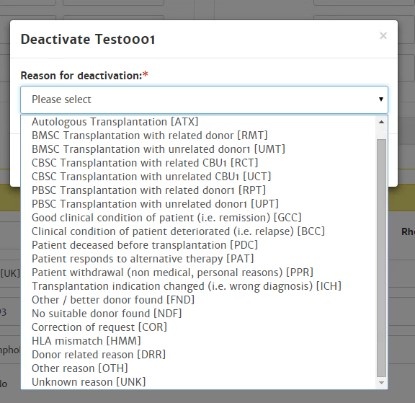
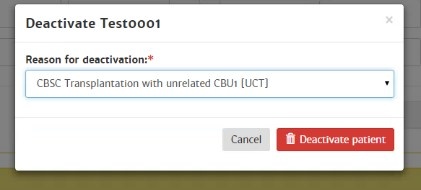
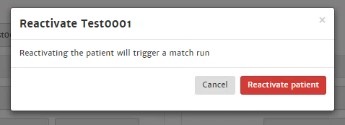
Deactivate patient
The system automatically deactivates your active patients when you have not performed any activities with this patient for more than 6 weeks (last viewed report date > 6 weeks). These patients will be archived in your inactive patient list with reason for deactivation: "system deactivated".
...
| Imagefloat | ||
|---|---|---|
| ||
Reactivate patient
Within the table of listed inactive patients you can manage your patients by performing the following actions listed below:
|
|
|
|
...
|
|
|
|
|
|
...
|
|
|
|
...
| Imagefloat | ||
|---|---|---|
| ||
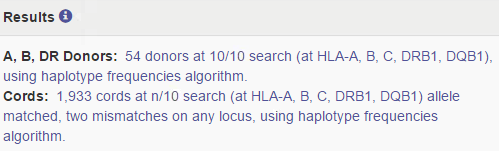
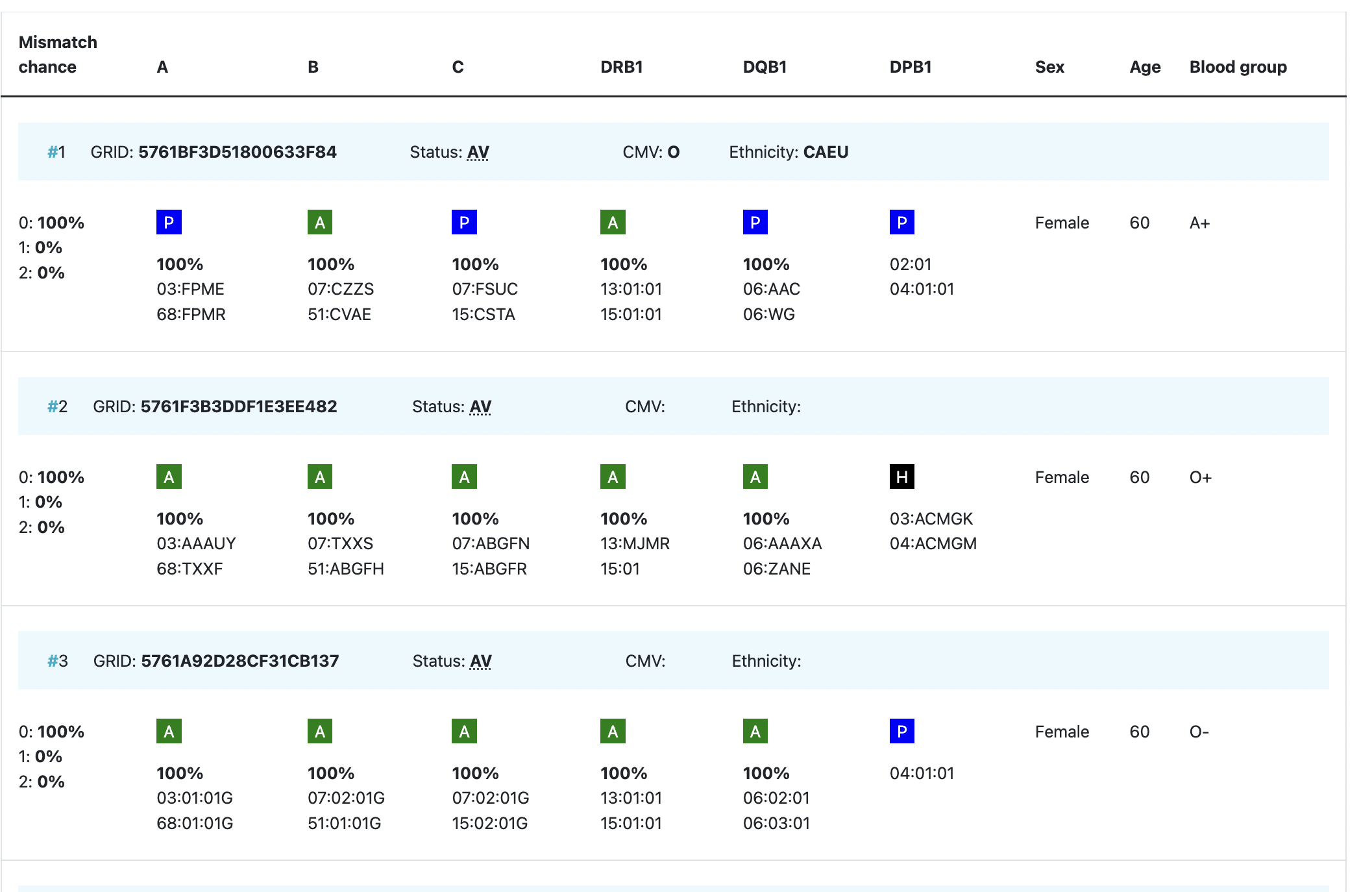
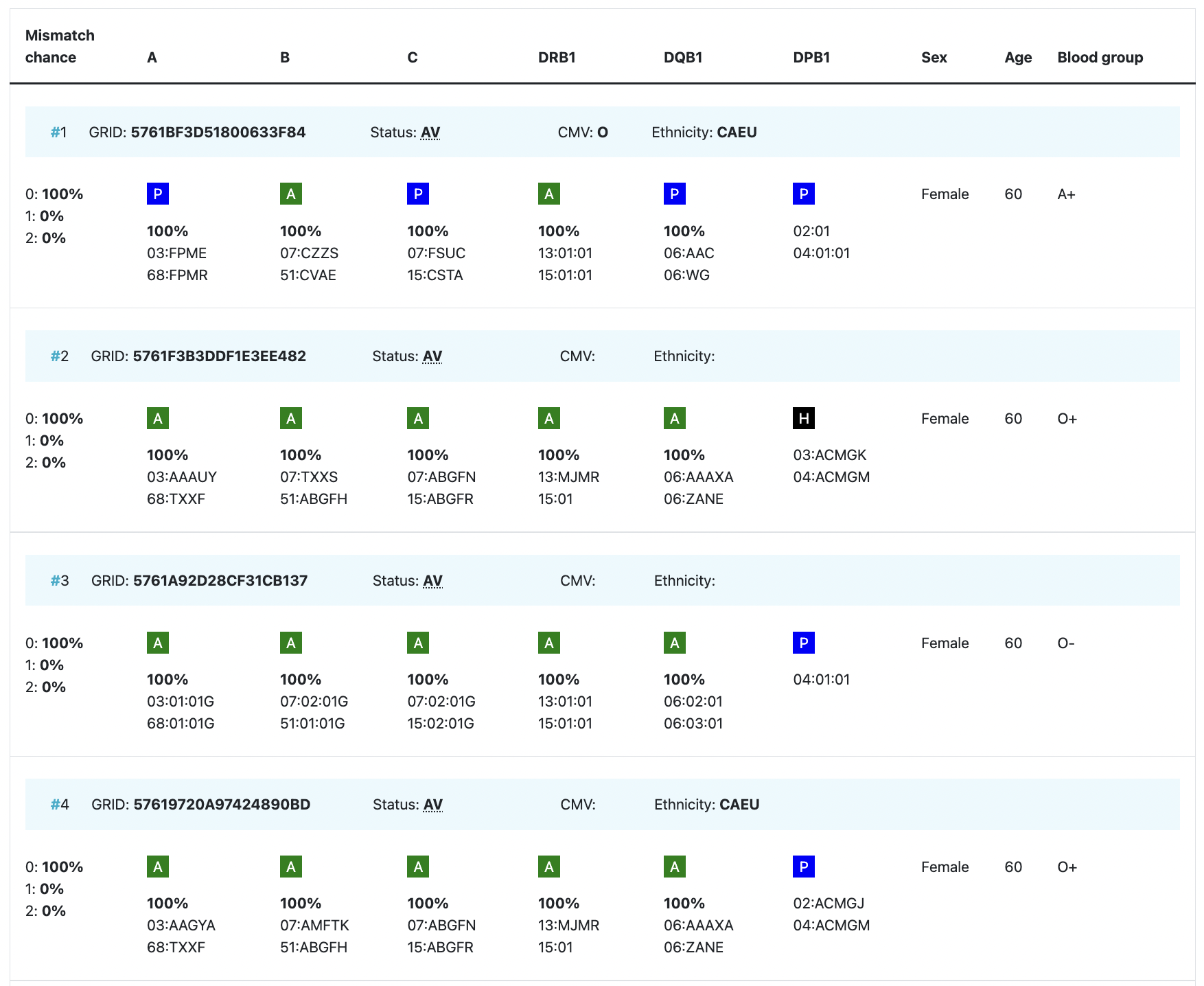
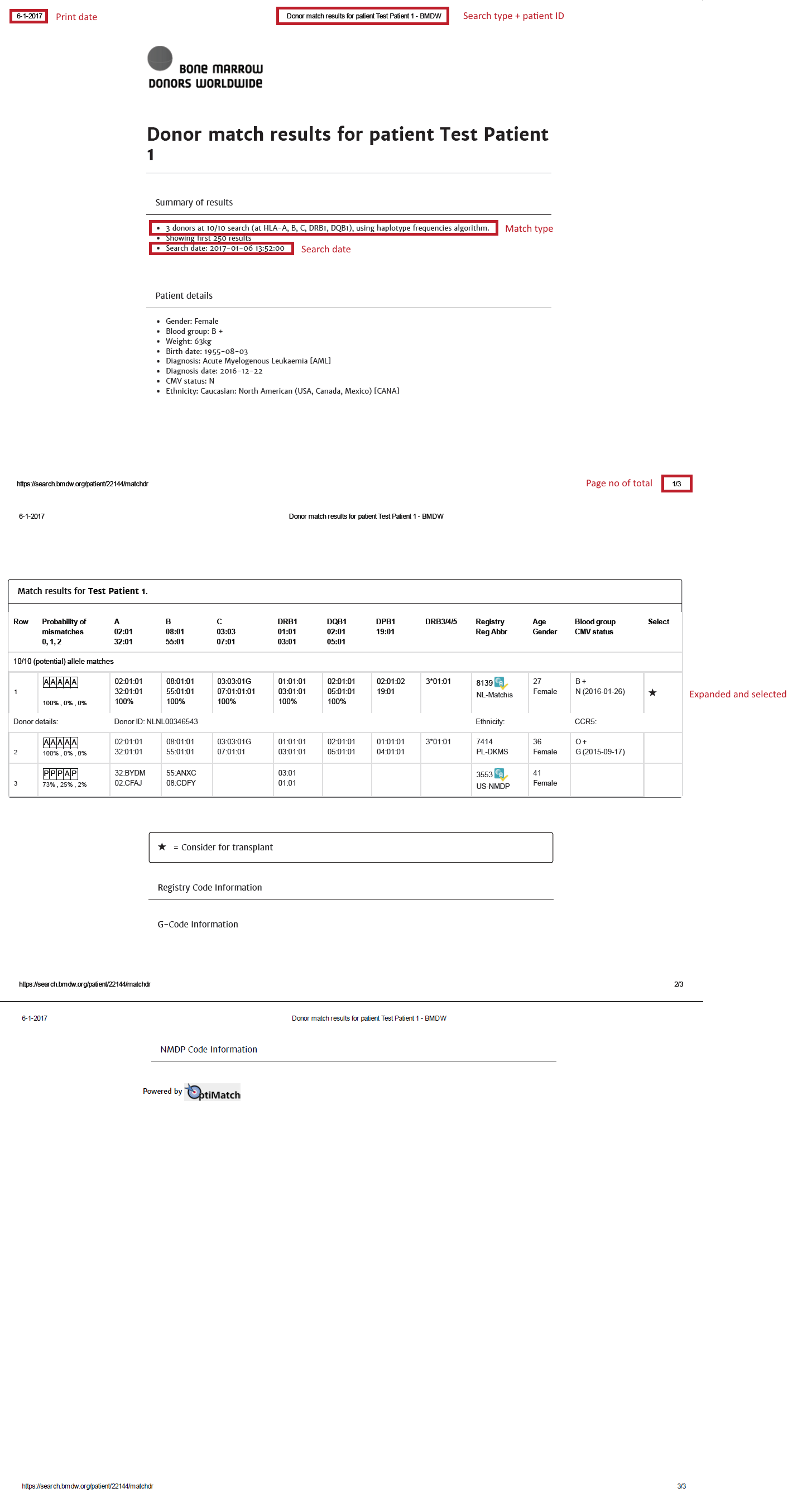
Overview of cords match results
| Imagefloat | ||
|---|---|---|
| ||
...
Abbreviation / column | Description |
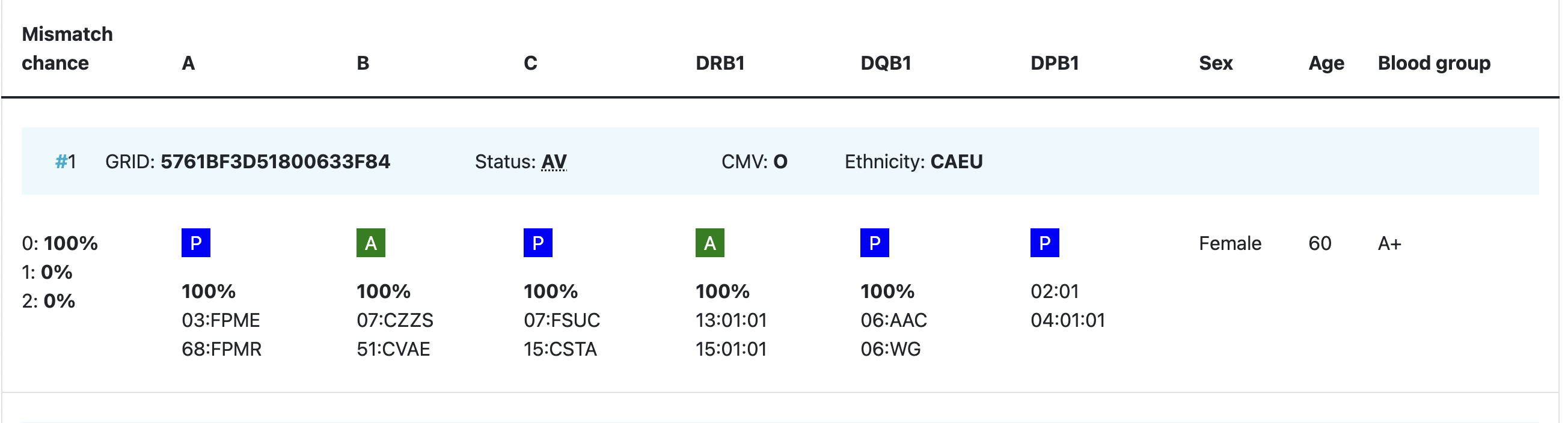
| HLA patient | In the grey bar, you can find the HLA of your patient. This header will move with you when you are looking at results more below.= |
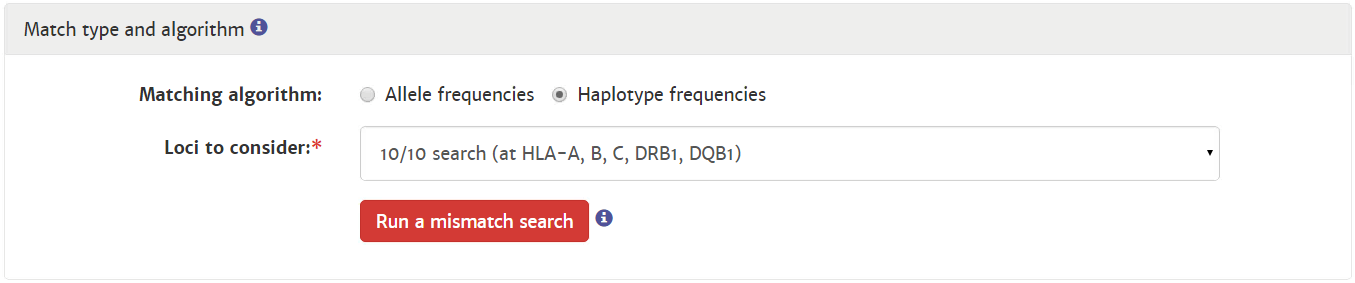
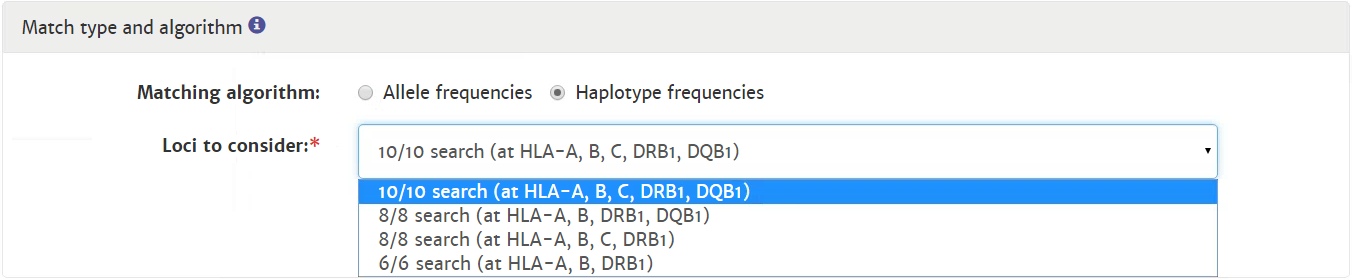
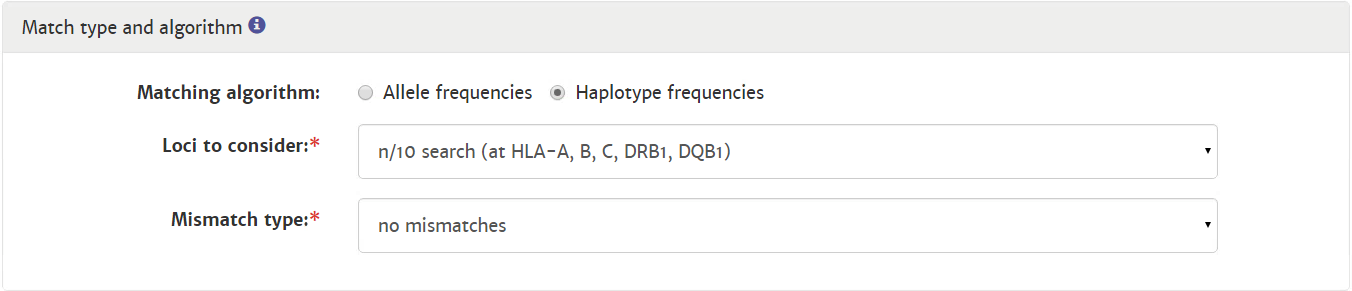
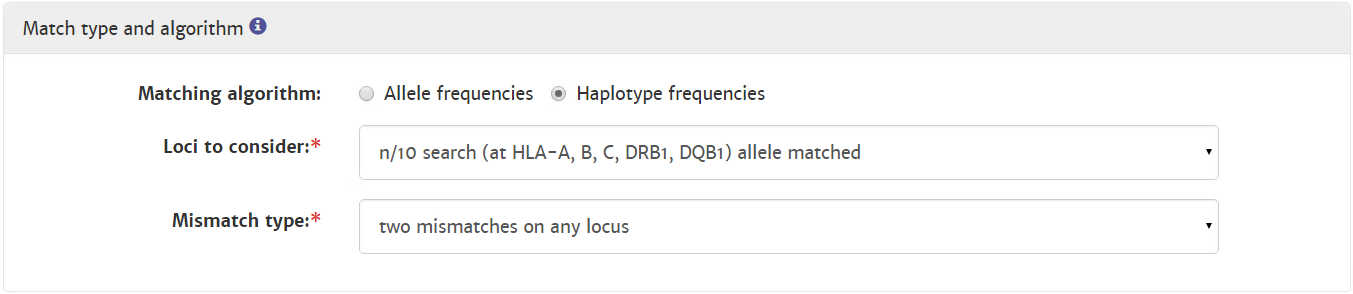
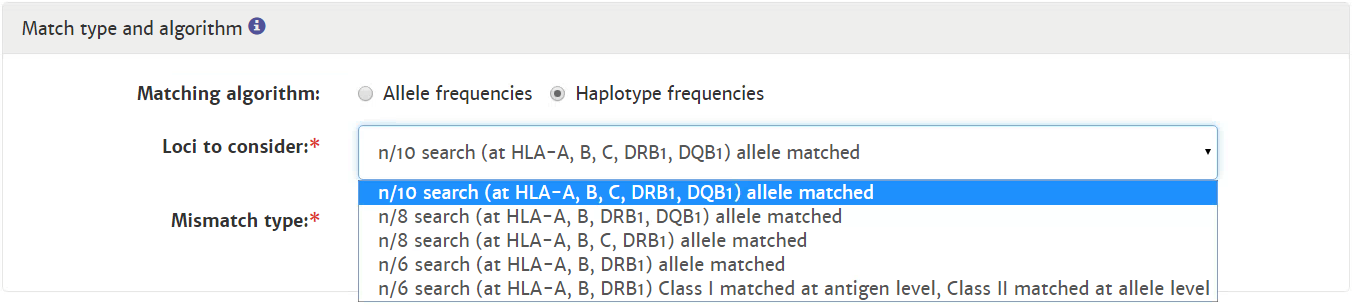
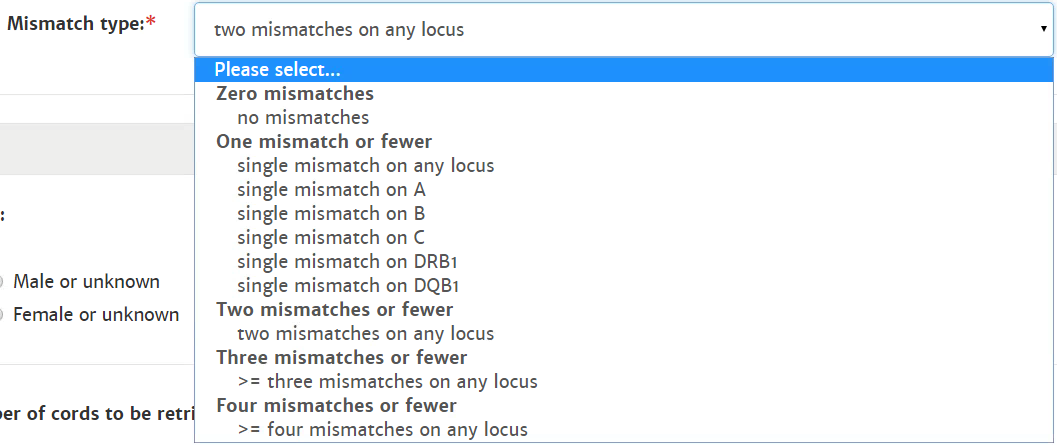
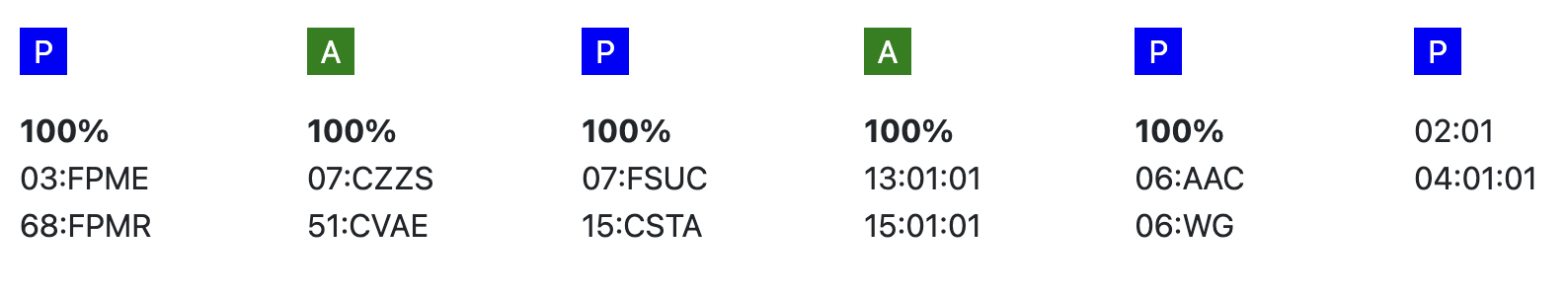
Probability of mismatches 0, 1, 2 | Probability of a mismatch at 0 loci, 1 locus, and 2 loci. The percentages are only shown when you are using the haplotype frequency algorithm and are based on the match type you have been chosen (out of 6 then 6 loci are considered; out of 10 then 10 loci are considered). The five squares above the probability percentages are representing, respectively, locus A, B, C, DRB1, and DQB1. They are showing in letter/colour codes if a certain loci of a donor/cord is likely to match with your patient or not.
NOTE: When you are using for cords the match type ≥4/6 (at HLA-A, B, DRB1), Class I matched at antigen level and Class II matched at allele level, the probabilities are calculated based on allele level match for all loci and not only for DRB1. |
| |
|
|
Age | Age of donor/cord |
Gender | Sex: M = male, F = female |
Blood group | Blood group, e.g. A+ = blood group A, rhesus positive, B- = blood group B, rhesus negative |
|
|
|
|
|
|
| Probability of match per locus | If you expand the donor/ cord details, the probability of a match per locus becomes visible: This also correspondents with the letter/colour code from the five squares in the column probability of mismatches. These probabilities are only calculated for the 5 loci A, B, C, DRB1, and DQB1. |
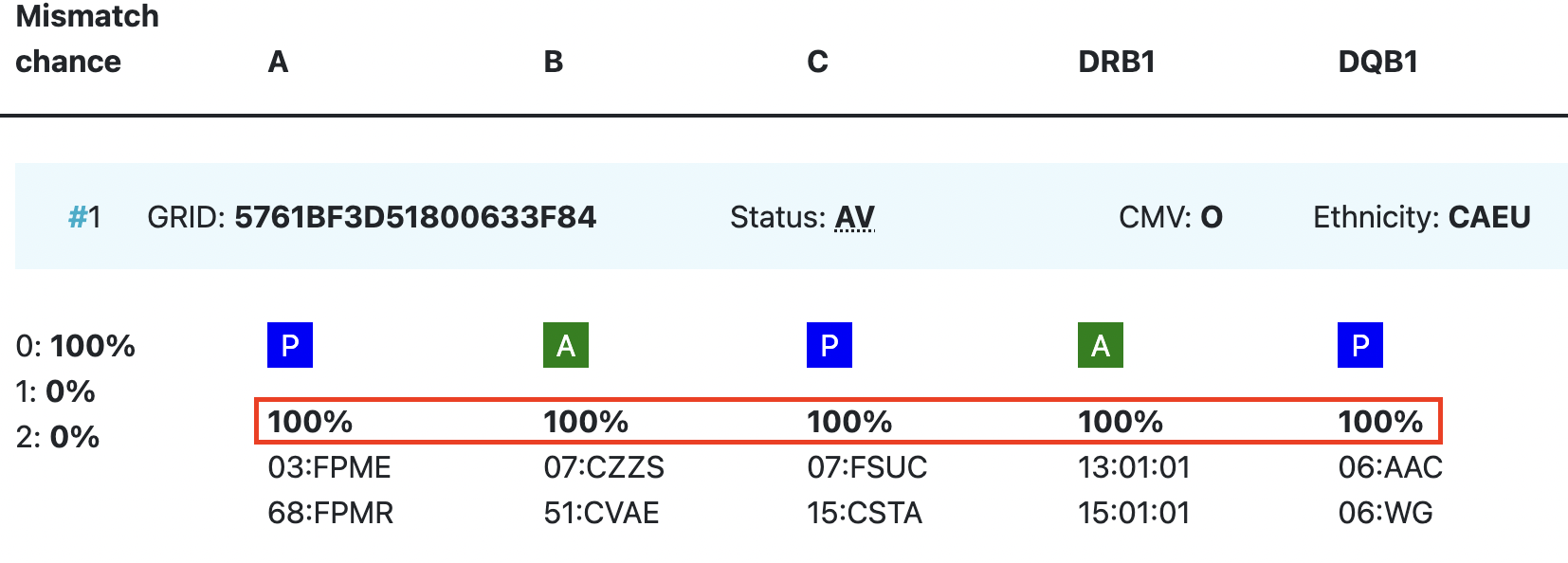
GRID, Global Registration Identifier for Donors, is a new ID for donors (not for CBUs) that is globally unique. The first 4 numbers of the GRID refer to the ION of the organisation where the donor is registered. From July 1st 2019, GRID will become the primary ID for communication purposes between organisations. | |
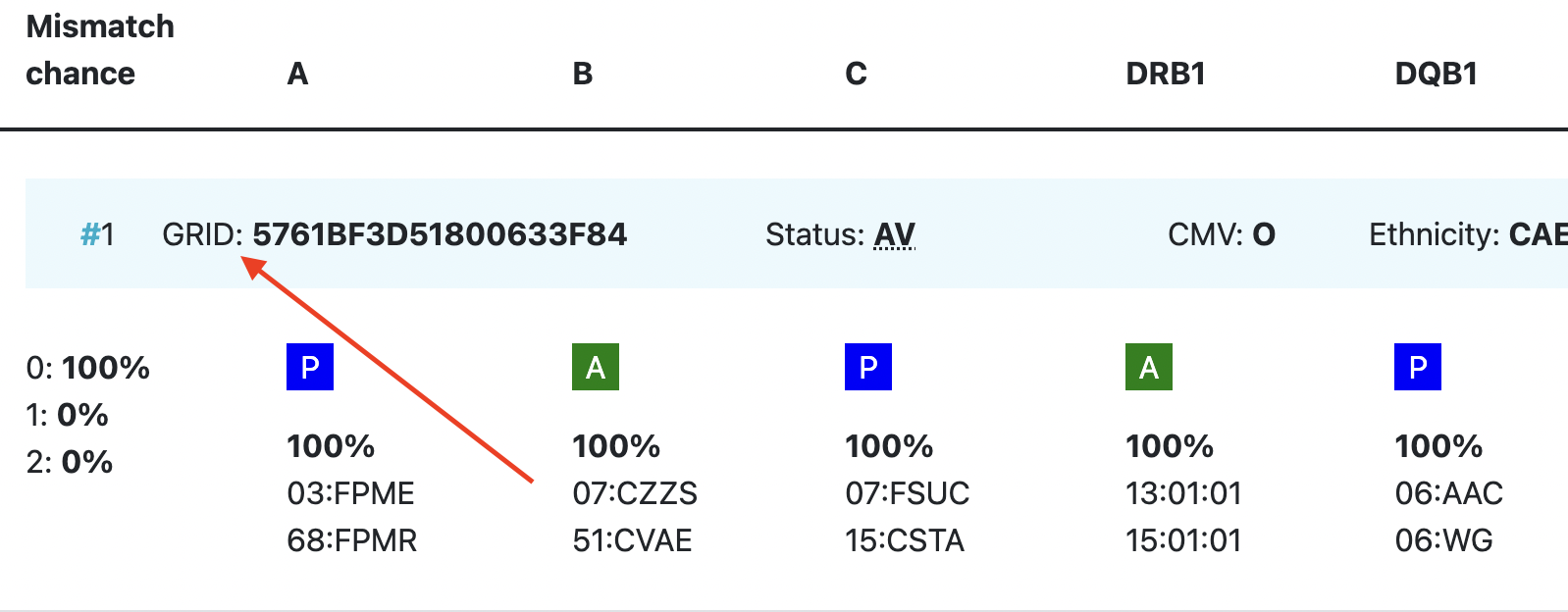
Ethnicity | Ethnic group: The system uses the same ethnic groups as defined for the EMDIS system: |
| |
|
...
| Imagefloat | ||||
|---|---|---|---|---|
| ||||
Footer
The footer section of the website contains links to various other useful information for you as a user (figure 23). All links with the icon , link out to another website, WMDA Share. Links with a icon, will open up an e-mail message. The report a problem link is only visible when you are logged into the website.
...