With the last release of our the Search & Match Service, the WMDA has maintained a similar haplotype frequency set configuration as was used in Optimatch (Search & Match v1). In most cases the ION of the donor/ cord blood unit will determine which haplotype frequency set is used.
To provide a more up-to-date representation of the donor pools world wide, the haplotype frequency set calculations have been performed using the latest data in the WMDA Donor and CBU database. Calcations were performed using the open source algorithm Haplo-o-Mat (https://github.com/DKMS/Hapl-o-Mat and https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5450239/) using high resolution typing results from organisations to extrapolate the haplotypes of the region or organisation. Thus, an organisation or geographical region must meet a minimum threshold of number of donors and availability of high resolution typing to build usable and valuable frequencies. For cord blood units. we will use the same sets that were determined in the donor populations.
The inclusion criteria aims to balance a high number of donors, the quality of the HLA types and the complexity of the haplotype frequency estimation. The number and HLA types of the loci that are included in the estimation can be independent of the loci of the haplotypes actually estimated if the quality of the former is appropriate.
5-locus Haplofrequency estimation was performed on all organisations/populations with at least 10000 donors in HLA-A, -B, -C, -DRB1 and -DQB1 types where ambiguity was low enough to be useful. Organisations in the same country/within the same population are usually combined, e.g. AT-ABMDR (ION-2614, Austrian Bone Marrow Donors) and AT-GFL (ION-4961, Verein Geben für Leben) but *not* for US-NMDP (general US population) and US-GOL (predominantly Ashkenazi Jews).
The current global frequency set will currently be used for all patients and for donors that for some reason do not match a typical haplotype frequency set. For example when a registry is new and has not yet been assigned to an appropriate existing haplotype frequency set or when the HLA typings from this registry deviate substantially enough from any typical haplotype frequency set that it does not make sense to assign them to one. The number of this frequency set is 999. We currently return the haplotype frequency set in the search results API endpoint for CBUs and will soon add it for donors.
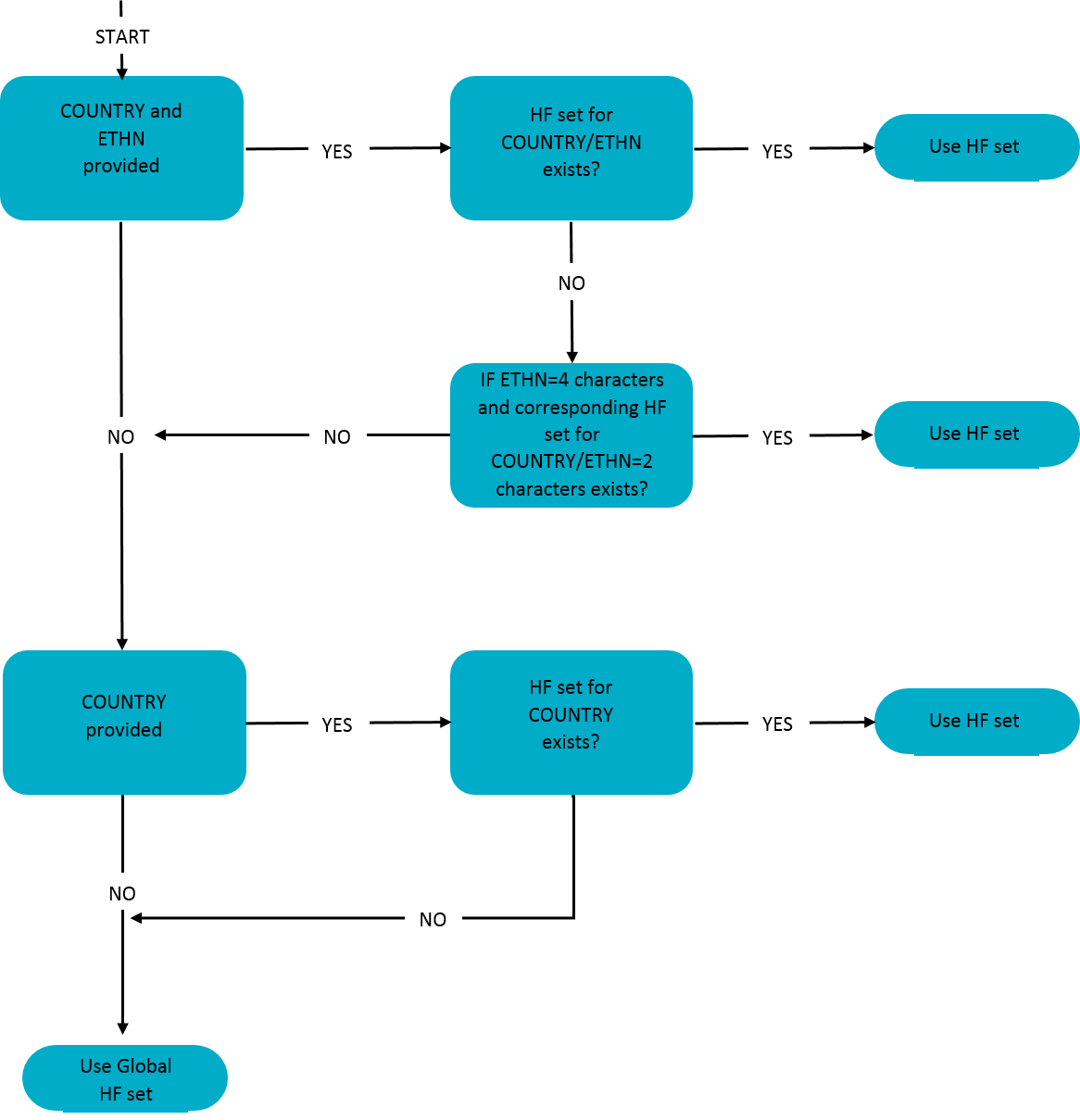
Below, you can find a figure how the haplotype frequency (HF) sets are assigned to the donors and cord blood units. The system first determines if both registry ION and ethnicity are provided. Registry ION is then used to look up the country/ region that it belongs to. If yes, then it will check if an ethnicity specific set for this registry exists. If the donor for example is coming from USA and has ethnicity HISA, then we do not have a specific set available. However, there is a more broad Hispanic HF set available (USA-HI) and this will then be used.
If there is no ethnicity available for the donor or no ethnicity specific HF set exists for that country, the system will check if a country specific HF set is available or if the country is part of one of the regional HF sets.
If no country specific HF set is available, the Search & Match Service will use the global consensus HF set.
Flowchart assigning a haplotype frequency (HF) set
In the table below, you can find the haplotype frequency set numbers and which countries/organisations were included as well as the total sample size. In the last part from the table we included regional sets, some country/ethnicity specific sets and the global consensus set.
The regional sets are defined as follows:
East Asia (eas): CN (CN+CN1), HK, TW, JP (Set number: 32)
Eastern Europe (eeu):AT (A+A2), CZ (CS+CS2), CY (CY+CY2), GR2, PL (PL3+PL5+PL6), TR (TRAN+TRIS+TRKK), BG, HU, HR, LT, MK, RU (R2+R4), RO, RS, SK, SI (Set number: 33)
South America (sam): AR, BR, UY (Set number: 34)
Input Data | Applied to: | |||||
HF set number(pop_id) | Determination date | Sample size | ISO country code | ION | Search & Match Service_Registry_codes | IONs |
| 1 | 2022-05 | 226507 | AR | 5117 | AR-INCUCAI | Same |
| 2 | 2022-05 | 149750 | AT | 2614 4961 | AT-ABMDR AT-Verein | Same + 8162 - AT-Vita34 (CBB) |
| 3 | 2022-05 | 41348 | AU + NZ | 7748 8261 | AU-ABMDR NZ-NZBMDR | Same |
| 4 | 2022-05 | 36351 | BE | 4201 | BE-MDPB | Same |
| 5 | 2022-05 | 81859 | BR | 8766 | BR-REDOME | Same |
| 6 | 2022-05 | 68077 | CA | 5103 6912 | CA-One Match CA-HemaQuec | Same + 3066 - CA-VAR (CBB) |
| 7 | 2022-05 | 114900 | CH | 9341 | CH-SBSC | Same |
| 8 | 2022-05 | 564150 | CN + HK + TW | 2197 6681 4070 3458 | CN-CMDP CN-Sunshine HK-HKBMDR TW-TzuChi | Same + 1212 - TW-SinoCell (CBB) 1714 - TW-Meribank (CBB) 3105 - HK-CBB (CBB) 5812 - TW-StemCyte (CBB) 6459 - TW-Bionet (CBB) 6692 - TW-Healthbanks (CBB) 9281 - HK-Mononuclear (CBB) |
| 9 | 2022-05 | 74633 | CY | 4278 9751 | CY-Paraskevaidio CY-CBMDR | Same |
| 10 | 2022-05 | 75810 | CZ | 4753 5440 | CZ-CSCR CZ-CNMDR | Same |
| 11 | 2022-05 | 2627544 | DE | 5525 6939 | DE-DKMS DE-ZKRD | Same + DE-DUS (CBB) All ethnicities except for ASSW |
| 12 | 2022-05 | 53849 | DK | 2015 7484 | DK-DSCDW DK-DSDE | Same |
| 13 | 2022-05 | 81265 | ES | 7813 | ES-REDMO | Same |
| 14 | 2022-05 | 39567 | FI | 9738 | FI-FSCR | Same |
| 15 | 2022-05 | 90069 | FR | 1804 | FR-FGM | Same |
| 16 | 2022-05 | 636595 | GB + IE | 6354 1726 2731 9968 5590 | GB-Anthony GB-WBMDR GB-BBMR GB-DKMS IE-IUBMR | Same |
| 17 | 2022-05 | 102358 | GR | 4979 | GR-HTO | Same |
| 18 | 2022-05 | 651085 | IL | 5239 4987 4068 | IL-Hadassah IL-Ezer Miz. IL-SHBB | Same |
| 19 | 2022-05 | 530272 | IN | 2824 4131 4460 8196 8486 9935 | IN-GeneBand IN-MDR IN-ArjanVir IN-ArjanVir IN-Datri IN-DKMS-BMST | Same |
| 20 | 2022-05 | 84477 | IT | 7450 | IT-IBMDR | Same |
| 21 | 2022-05 | 328557 | NL | 8139 | NL-Matchis | Same |
| 22 | 2022-05 | 16499 | NO | 7214 | NO-NBMDR | Same |
| 23 | 2022-05 | 1464544 | PL | 3918 5391 7414 | PL-ALF PL-Poltranspl PL-DKMS | Same |
| 24 | 2022-05 | 11029 | PT | 7358 | PT-Cedace | Same |
| 25 | 2022-05 | 97139 | SA | 1810 2107 | SA-KFSHRC SA-SSCDR | Same |
| 26 | 2022-05 | 184283 | SE | 5285 | SE-Tobias | Same |
| 27 | 2022-05 | 78885 | SG | 3785 | SG-BMDP | Same + 4291 - SG-SCBB (CBB) |
| 28 | 2022-05 | 122133 | TH | 8362 | TH-TSCDR | Same |
| 29 | 2022-05 | 491941 | TR | 3893 5509 3503 | TR-TRAN TR-TRIS TR-TURKOK | Same |
| 30 | 2022-05 | 1723292 | US | 3553 | US-NMDP | 3553 with ethnicities:
and the following organisations (all ethnicities) 4857 - US-CSCC (CBB) 6579 - US-Cleveland (CBB) 6738 - ZA-DKMS 7470 - US-Lifebank 8118 - ZA-SABMR 8379 - US-StemCyte (CBB) 8691 - US-NCBP (CBB) |
| 31 | 2022-05 | 97953 | US-GOL | 1033 | US-GOL | Same |
| 32 | 2022-05 | 564561 | CN+HK+TW+JP | 1212 1714 2197 3105 3458 4070 5812 6459 6681 6692 6933 | 4364 - JP-JMDP 8405 - KR-KONOS All other IONs that were used as input have their own country/region specific set. | |
| 33 | 2022-05 | 2398213 | AT+CZ+CY+GR+PL+TR+BG+HU+ HR+LT+MK+RU+RO+RS+SK+SI | 1005 1372 1695 2073 2614 3503 3893 3918 4278 4307 4398 4565 4650 4753 4961 4979 5019 5391 5440 5509 5712 7197 7414 8162 8256 8714 9751 9778 | 1005 1372 4381 4398 4565 4650 5712 7197 8256 8714 9778 | |
| 34 | 2022-05 | 413438 | AR+BR+UY+CL+PY | 1574 2547 5117 6517 8766 | CL-DKMS PY-VKS AR-INCUCAI UY-SINDOME BR-REDOME | 1574 - CL-DKMS 2547 - PY-VKS 4675 - CL-Vidacel (CBB) 6517 - UY-SINDOME Brazil and Argentina have their own country-specific sets. |
| 35 | 2022-05 | 38530 | ZA | 6738 8118 | ZA-DKMS ZA-SABMR | None yet. Under investigation. |
| 36 | 2022-05 | 35895 | IR | 4993 6887 | IR-INSCDN IR-ISCDP | None yet. Under investigation. |
| 41 | 2022-05 | 11012 | LU | 3099 | LU-LMDP | Same |
| 100 | 2022-05 | 151,204 | DE-ASSW | 5525 6939 | Subset of donors from these registries with ethnicity:
| Same |
| 101 | 2022-05 | 656,591 | US-AF* | 3553 | Subset of donors from this registry with ethnicities:
| Same |
| 102 | 2022-05 | 796,780 | US-AS* | 3553 | Subset of donors from this registry with ethnicities:
| Same |
| 103 | 2022-05 | 3,740,668 | US-CA* | 3553 | Subset of donors from this registry with ethnicities:
| Same |
| 104 | 2022-05 | 1,002,893 | US-HI* | 3553 | Subset of donors from this registry with ethnicities:
| Same |
| 999 | 2022-05 | 11,430,561 | All donors | All donors/ CBUs not specifically matching any other set. | ||
Since WMDA is doing these calculations on behalf of organisations, it is important for organisations to submit high resolution typing when available and donor ethnicity data. This greatly improves the haplofrequency sets generated and allows for individual donor match grades to be more accurate.
NOTE: None of the Japanese and Korean donors have been typed for DQB1. This means that no Japanese or Korean donors were included in the calculations for set #32. This means that the haplotype frequency set applied to Japanese and Korean donors are probably biased against Japanese and Korean haplotype frequencies. We have kindly received a self-generated haplotype frequency set from the Japanese registry and are working on integrating it in the Hap-E algorithm that is part of Search and Match. Whether to apply the provided Japanese frequency set or to apply set #32 is still under investigation.
NOTE: in some cases a countries donors may be used in a national haplotype frequency set as well as a regional one. For example Austrian donors are used in their own set (#2) as well as the Eastern Europe region set (#33). When calculating match probabilities always the most specific set is used. So for donors from IONs 2614 or 4961 set #2 is used, but for Slovakian donors there were not enough high resolution donors for their own set so the regional set is used (#33).
There are also countries or donors with a specific ethnic background in a country for which there was no specific haplotype frequency set in Optimas, but for which there are now enough donors for their own haplotype frequency set and/ or their haplotype . We are currently investigating whether it would provide benefit to host these separately. These include:
- IN-DATRI. Since DATRI is recruiting in a specific region of India, its haplotype frequencies may be significantly different from the general Indian frequencies (set #19)
- ZA: there are enough donors to merit its own set. Considering the unique make-up of the ethnic background of South African donors, having frequency sets per ethnic group would be especially beneficial.
It is important to know that your organisations frequency set is changeable!
You can request that your donors be returned to the global consensus haplofrequency set or to a regional set. Your request should include the reasons why you believe that another set is more applicable for your donors. Additionally, if your organisation has a frequency set available that you would like to see utilized for the donors of your organisation we can implement it as well. In this case, your request should include at least a description about your population, the sample size, inclusion and exclusion criteria, calculation method, and the reasons why your haplofrequency set would be better than the current set applied by WMDA. Both requests will be reviewed by the WMDA BioInformatics Working Group. Please send requests to support@wmda.info . In the future, you'll be able to dictate a frequency set on a per donor basis.
| Date | Version | Description | Author |
|---|---|---|---|
| 2017-10-31 | 1.0 | Replacement BMDW global haplotype frequency set for more specific sets | JK |
| 2018-01-24 | 1.1 | Modification some sets; introduced during OptiMatch version 3.31.0 | JK |
| 2019-02-15 | 1.2 | Replaced BMDW by WMDA / Search & Match Service; updated email address | JK |
| 2022-08-15 | 1.3 | Copied original page and updated with relevant info for HAP-E algorithm. | MM |