- Created by Mark Melchers, last modified by Alicia Venter on Jan 19, 2023
You are viewing an old version of this page. View the current version.
Compare with Current View Page History
« Previous Version 40 Next »
Questions are divided into the following subjects:
GENERAL
Requesting authorization:
The matching programmes are only available for registry staff members and staff members of organisations assisting in searches for unrelated transplants. Find below the procedure to get access for the different types of users. You can register on the WMDA website.
For registered users, multi-factor authentication has been applied as of 2022. Please send an email to support@wmda.info for credentials to perform your initial authentication.
WMDA provides search advise for difficult cases. You can request a search advise at the bottom of the Update patient screen and the HLA experts will then help you to determine the best search strategy for your patient.
PATIENT MANAGEMENT
No. Because of the way the two systems are built they do not share the same database. Therefore patients from V1 will not be available in V2 by default. If you'd still like to have your V1 patients visible in the V2 system, there are three ways to achieve this:
- You implement the patient API endpoints for the V2 system and use it to send over the patients in V1 to V2. This could also be just the relevant patients. You could then keep using the patient API V2 implementation to send patients from your local system to Search & Match V2.
- You provide us with a list of requirements for patients in your V1 system and we will port them over to the new system. The WMDA will only do this once for an organisation.
- You manually enter the relevant patients from the V1 system into the V2 system.
Patient information can be accessed from the 'Patient List' by selecting the "Patient ID'.
Patients for whom a search was performed within the last 6 weeks will be in the ‘Active patient’ list, while the previous 5 months will be in the ‘Inactive patient’ list.
If you cannot find your patient within the default Active list remember to search the Inactive list also.
Users will have access to patients registered by other users at their organisation as well, which is also split into 'Active Patients' and 'Inactive patients'
No. De-activating a patient will just remove the searches and search results from the patient and place the patient in the 'Inactive patients' list.
If you need to re-activate a patient, from the Patient List section open the 'Inactive patients' list and search for the relevant patient. On the far right of your screen, in line with the de-activated patient's information, you will find a button that reads "Reactivate", which you need to click.
When you open a patient record from your inactive patients, the details cannot be edited. If you need to change details like weight, you first have to reactivate the patient. When the record is placed in the Active patient list again, you can update the details of your patient.
The patients in the patient list are default sorted on the date of last viewed. Records that have been created are also considered to have been "viewed" and there will therefore appear at the top after creation.
The easiest way is to change the view first in your "Active" patient list and then go to the Inactive patient list.
No, the system does not auto correct HLA information, but may give you error warnings as applicable.
SEARCHING
Hap-E searches may return donors that do not have typing at DRB1, but do have typing at loci A, B and at least one other locus. Since these donors are usually not considered relevant potential donors and there tend to be quite a few of these donors that potentially match the patient, Search & Match applies a default filter which removes all donors that do not have typing at DRB1. You can see this by the little "1" next to the filter.
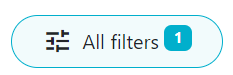
This means that there is already a filter applied to the results.
If you are interested in the donors without DRB1 typing you can remove the filter and see all results.
If this occurs the most likely cause is:
- Your patient has homozygous typing at at least 1 locus
- The potential donor/CBU has low resolution typing at this locus or a MAC code that contains a null allele
The reason this donor/CBU is shown as a potential match and not a mismatch is that the typing for the donor contains a null allele. It is therefore a possibility that the donor actually has a null allele. Since a homozygous typing is generally considered a match with a donor that has one allele that matches the homozygous patient typing and the other is a null allele, Hap-E returns this donor/CBU as a potential match. Please see the slide below from a recent webinar:
In the legacy system when running a standard donor search, all returned donors have values for A, B and DRB1. This is different when running a search in the new system. There all donors need to have typing at A, B and at least one other locus (which may or may not be DRB1). This means that donors are returned in the new system that do not have typing at DRB1. This typically explains the majority of the difference in number of search results as mentioned in the search summary.
Although in some cases these donors with typing at A, B and C or DQB1 may prove useful, in most cases they are not. We therefore apply a default filter to all search results.
You can see all donors by unchecking the default filter.

Because the filter is applied by default, you may notice a difference in the total number of donors/cords as described in the search summary text versus the number displayed just above the list of potential matches. For example in the example below, there are 33 donors in the full search results. 3 donors did not have typing at DRB1 and have therefore been filtered out in the donor search results shown.
You can see that a filter has been applied by the "1" icon on the "All filters" button.
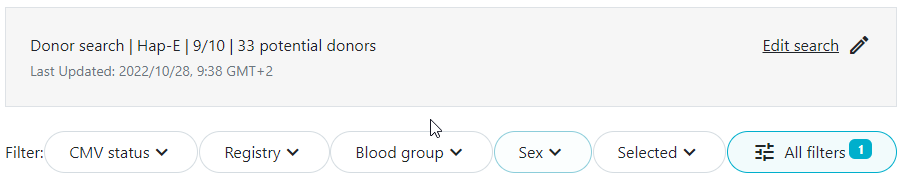
Asynchronous matching is a process of running searches in parallel. It allows you to enter a patient and start the search run, and proceed with other activities within the application like entering another patient or reviewing another match list while the search runs in the background. You do not have to wait for the current search to finish before proceeding with other activities.
Many factors can impact how long it takes to retrieve search results. The most likely reasons that your search is taking some time is that it is a difficult search or the HLA of your patient is very common and will retrieve many records. We initially only run a 10/10, 8/8 or 6/6 search to make sure we retrieve you results as quickly as possible. Mismatch searches can take longer since there are many more options to assess before providing the results.
By default, Search & Match performs a 10/10 search for donors if all 5 loci (A,B,C,DRB1,DQB1) are available for the patient. If only 3 or 4 loci are available, the system will scale down to a 6/6 or 8/8 search, respectively. 10/10, 8/8 and 6/6 are all considered as zero mismatch searches and will not include mismatch donors in the results. This is also the case for cord searches.
Open up the search results for a search you would like to perform a mismatch search. Click "Edit search" and then select the number of mismatches that you would like to consider at most. Then click "Search" to start that search.
No, you can only perform a zero mismatch donor or cords search. You will only be able to view/request mismatched donors/CBUs when the initial search results are available.
If you are experiencing difficulties in your selection of a donor/cord for your patient, you can request a search advice from the WMDA HLA experts. To request an advise, go to your patient list, click on the patient ID and at the bottom of the Update patient form you will find a button with 'Request Search Advisory'. An email message will then open with the patient information and some additional fields with requirements. Please fill out as much as possible and send the email. You will receive the advice also by email.
There is no functional difference. The filters you see on top of the headings in the search results are the frequently used filters. All filters are applied to all search results.
The default sorting criteria from Hap-E Search and ATLAS for donors are:
1. HLA
2. probability in 10% intervals
3. donor age ascending
The default sorting criteria from Hap-E Search and ATLAS for cords are:
1. HLA (6/6, 5/6, 4/6 categories)
2. Number of total nucleated cells (TNC) for cords within HLA match category
DPB1 TCE3 evaluation is performed only for A-B-DR(B1) typed donors under the following conditions:
- Patient DPB1 values must be present. Typing ambiguities in the form of multiple alleles codes, G-codes, etc. are allowed.
- Donor DPB1 values must be present. Typing ambiguities in the form of multiple alleles codes, G-codes, etc. are allowed.
As part of the DPB1 TC3 grading filter, you can filter the results by the following options: Matched, Permissive, Non-permissive in HvG direction, Non-permissive in GvH direction, Ambiguous/Undetermined.
Although the DPB1 TC3 filter is available for cords, the results haven't been validated so it shouldn't be used.
The default sorting criteria from Hap-E Search and ATLAS are:
1. HLA
2. probability in 10% intervals
3. donor age in 5 year intervals
This can have several causes.
- Overall match probabilities and locus-specific match probabilities are missing: this is most likely because the HLA typing of the donor/CBU is not consistent with any known haplotypes for the population that the donor is in (inexplicable donor). Without haplotypical context, the matching engine is not able to provide match probabilities.
- Only locus-specific match probabilities are missing: It does not make sense to display the probability of match for a specific locus in case of a mismatch
- Because the known match probability is 100% and therefore there is no probability of a(n additional) mismatch and therefore displaying match probabilities in case of an additional mismatch does not make sense.
- Because the probability of an additional mismatch is 0% for another reason, e.g. because the donor is low resolution typed at 3 loci and therefore in theory be a match, but it is very unlikely.
See the following page for more information: Feature differences Hap-E Search vs Optimatch
At the top of search results you can see how many donors or CBUs are inexplicable. 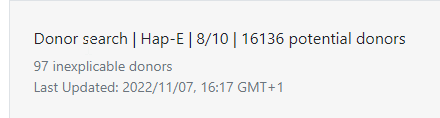
Inexplicable donors or CBUs are records that have HLA that cannot be explained by a combination of two haplotypes in the haplotype frequency set (Haplotype frequency sets in the HAP-E matching algorithm of the Search & Match Service) for the population that the donor or CBU is part of. To find out which haplotype frequency set was used, you can use the full report
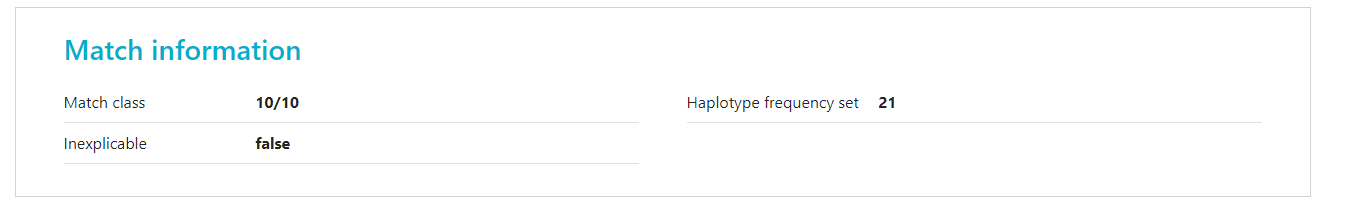
The number there corresponds to the set on the Haplotype frequency sets in the HAP-E matching algorithm of the Search & Match Service share page.
Because the HLA typing that this donor/CBU has cannot be explained, Hap-E also cannot calculate match probabilities. Currently these donors appear on the bottom of the match class that they are part of (e.g. 10/10, 9/10, 7/8). We are working on a way to move potentially relevant but inexplicable donors to a better place in the search results.
The number of inexplicable donors/CBUs mentioned therefore serves as a reminder that there are potentially relevant search results available that can be found at the bottom of the match class.
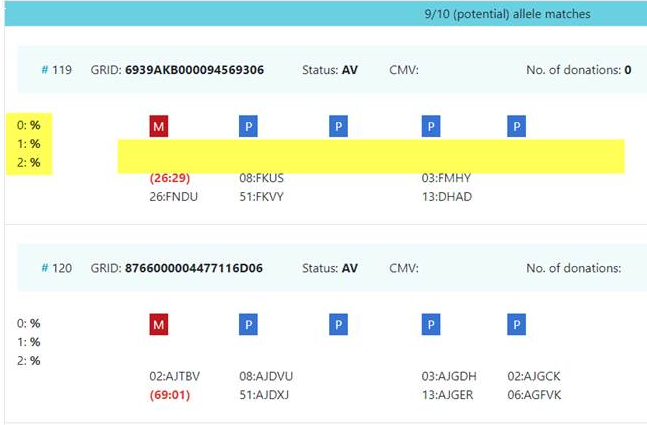
Since B*15:BPXE = B*15:03/61/74/103 a donor with this codes is a potential allele match for the patient. According to the official WMDA serology/DNA correspondence table, B*15:61 and B*15:74 have a serology of B15/B70 while B*15:03 is B72 and B*15:103 is B70. As a consequence a serology of B15 rules out B*15:03 and this donor is no longer potentially identical (on the allele level). Another explanation could be the limited length of donor lists.
Background: Unfortunately, for many donors serology was derived from DNA (by using the first field for the serological assignment) and vice versa (by appending “:XX” to the serological assignment) and often eventually both values are reported. In the case discussed, B*15:BPXE probably was translated back into B15 which is most likely wrong. This is a typical and (with certain registries) frequent case but there are many more unexpected DNA-serology correspondences that can give rise to exactly the same situation.
Search and Match does not consider binary attributes like gender or CMV for sorting at all. If you see a rich donor list and prefer, for example, male donors you should apply the filter to see male donors only.
Rationale: There is no agreed concept for weighing secondary match criteria like age, gender or CMV against each other. The approach to sort by probability in blocks and then by age plus ad hoc filtering gives the user maximum control of the appearance of the list.
This can happen only if the difference in resolution is not considered sufficiently relevant in the given context by Hap-E Search and ATLAS. Below are some possible (not mutually exclusive) reasons:
- The allele designations encode for the identical ARD (Antigen Recognition Domain) and Hap-E Search and ATLAS only matches for the ARD part of the HLA protein complex. For example B*07:ANVB stands for B*07:02/07:61 which both encode for the same ARD (here: exon 2 and 3). Hence Hap-E Search and ATLAS is regarding the codes B*07:02, B*07:61 and B*07:ANVB all as allele matches for each other but might give different matching probabilities for phenotypes containing each of those three codes since haplotypes involving B*07:02 and B*07:61 are having individual frequencies.
- The resolution only looks higher but, in fact, is lower (e.g. B*15:14 currently is only a single allele while B*15:14:01G covers three), identical (e.g. A*80:01, A*80:01:01 and A*80:01:01G cover exactly the same alleles) or not directly comparable since each code is containing some alleles not covered by the other (e.g. comparing B*07:TDVB and B*07:02:01G the former also covers a lot more variants of B*07:02 like B*07:02:02 to B*07:02:05 while it is excluding other alleles like B*07:44 and B*07:49N).
- The broader code only contains additional alleles which have never been observed in the population whose haplotype frequencies are used - at least not in a relevant haplotypical context. Their frequency could also be so low that it is disregarded due to rounding or the 10% probability grouping explained elsewhere. This applies, in particular, to all null variants of expressed alleles with identical first two field designations.
CASE STUDIES
- If the HLA of the patient does not contain information for all 5 loci, the system will automatically scale the match run down to 8/8 or 6/6 depending on the available HLA information of the patient.
This typically happens for donors when a donor's phenotype cannot be explained by the haplotypes used for probabilistic matching. It is therefore impossible to calculate match probabilities both at the considered loci in total and per-locus.
An 8/8 search does not consider DQB1 and therefore will not show whether a donor has or does not have a mismatch for DQB1. If the donor only has a known mismatch at DQB1, this donor will show up in the search results as a potential 8/8 match.
A*02:01 (2) vs. 02:03 (203) is an allele mismatch because A203 is an associated antigen to A2.
B*15:01 (62) vs. B15:03 (72) is an antigen mismatch.
The probabilities shown in your match results table are based on allele level matching on all loci.
Currently this feature is not implemented. However, this could be a feature for upcoming versions of the matching program.
- No labels