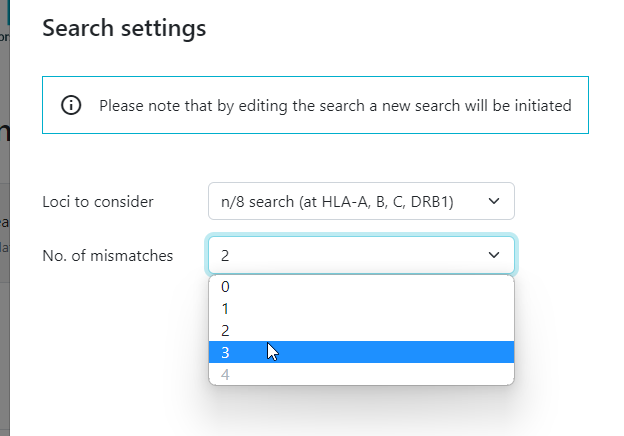
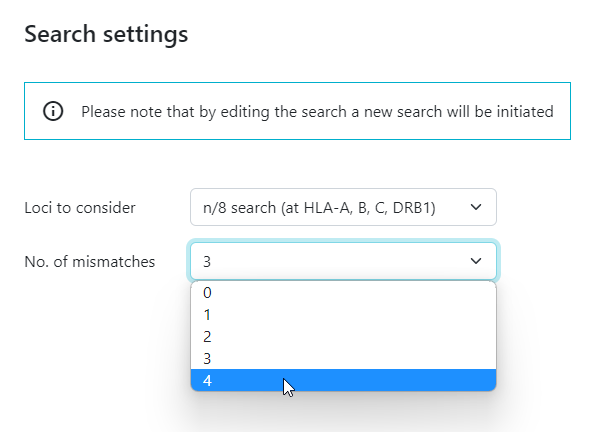
3 and 4 mismatch searches have now been implemented for Cord searches. Here are some things you need to know:
- In order to minimise unnecessary use of resource-intensive 3 and 4 mismatch cord searches, you first need to have finished
- a 2 mismatch CBU search before you can start a 3 mismatch search
- a 3 mismatch CBU search before you can start a 4 mismatch search
- a 3 mismatch CBU search before you can start a 4 mismatch search
- a 2 mismatch CBU search before you can start a 3 mismatch search
- Just like in the legacy system, no overall match probabilities are calculated and sorting is performed within the match class (i.e. 5/8. 4/8) based on the TNC count or CD34 count (depending on what you have selected. TNC count is the default)
- The number of search results can be bigger than it was when using the legacy system. It is advisable to use filters as the number of results can be great. You can for example filter on the minimum TNC, minimum CD34 or by certain registries or mismatch locations.
Future improvements
Score based sorting of 3 & 4 mismatch search results
As mentioned above, since 3 and 4 mismatch CBU search results do not have any match probabilities calculated, the results are sorted by TNC or CD34. The Hap-E algorithm in the new Search & Match service returns more potential CBUs than the legacy system. Many of these “extra” CBUs tend to be low probability CBUs that have low resolution or missing typing. This is not actually “wrong” because they are a potential match, but they tend to not be good candidate CBUs.
Because match probabilities are currently not calculated, sorting is done by TNC or CD34 only à poorly matched CBUs with high cell dose are at the top of the list and potentially better CBUs with a high enough cell dose are lower
Already planned ways of dealing with this
- Filter for returning only donors/CBU with typing at C (and/or DQB1) à in progress
- Filter on match class so you can go directly to the 5/8 or 4/8 potential matches à in progress
New method, soon to be available in sandbox:
- Assign a score based on the typed loci and their resolution
- Each locus can have one of the following “resolutions”
- High resolution typing (only one ARD)
- Low or intermediate typing
- No typing
- Each resolution class per locus has a “score”. Proposal:
- High = 20
- Low or intermediate = 17
- No typing = 0
- We sort CBUs based on the sum of the score first and only then by TNC/CD34
- n/6: sum of score at A, B, DRB1 (also at C?)
- n/8: sum of score at A, B, C, DRB1
- n/10: sum of score at A, B, C, DRB1 and DQB1
- Sorting for 0 , 1 and 2 mismatches remains unchanged